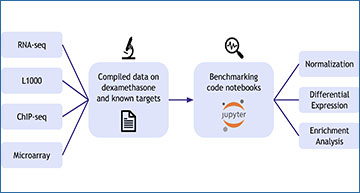
Many tools and algorithms are available for analyzing transcriptomics data. These include algorithms for performing sequence alignment, data normalization and imputation, clustering, identifying differentially expressed genes, and performing gene set enrichment analysis. To make the best choice about which tools to use, objective benchmarks can be developed to compare the quality of different algorithms to extract biological knowledge maximally and accurately from these data. The Dexamethasone Benchmark (Dex-Benchmark) resource aims to fill this need by providing the community with datasets and code templates for benchmarking different gene expression analysis tools and algorithms. The resource provides access to a collection of curated RNA-seq, L1000, and ChIP-seq data from dexamethasone treatment as well as genetic perturbations of its known targets. In addition, the website provides Jupyter Notebooks that use these pre-processed curated datasets to demonstrate how to benchmark the different steps in gene expression analysis. By comparing two independent data sources and data types with some expected concordance, we can assess which tools and algorithms best recover such associations. To demonstrate the usefulness of the resource for discovering novel drug targets, researchers at the Icahn School of Medicine at Mount Sinai applied it to optimize data processing strategies for the chemical perturbations and CRISPR single gene knockouts from the L1000 transcriptomics data from the Library of Integrated Network Cellular Signatures (LINCS) program, with a focus on understudied proteins from the Illuminating the Druggable Genome (IDG) program. Overall, the Dex-Benchmark resource can be utilized to assess the quality of transcriptomics and other related bioinformatics data analysis workflows.
The Dex-Benchmark Resource provides datasets and template workflows needed for benchmarking different tools for gene expression analysis. These datasets include microarray, RNA-seq, L1000, and ChIP-seq data related to dexamethasone and known targets such as NR3C1. Template workflows can be downloaded along with raw data to benchmark a set of gene expression profiles. Example code and visualizations are also provided.
Availability – The resource is available from: https://maayanlab.github.io/dex-benchmark.
Xie Z, Chen C, Ma’ayan A. (2023) Dex-Benchmark: datasets and code to evaluate algorithms for transcriptomics data analysis. PeerJ 11:e16351. [article]