Adapter trimming is an essential step for analyzing small RNA sequencing data, where reads are generally longer than target RNAs ranging from 18 to 30 bp. Most adapter trimming tools require adapter information as input. However, adapter information is hard to access, specified incorrectly, or not provided with publicly available datasets, hampering their reproducibility and reusability. Manual identification of adapter patterns from raw reads is labor-intensive and error-prone. Moreover, the use of randomized adapters to reduce ligation biases during library preparation makes adapter detection even more challenging.
Researchers at the Vanderbilt University Medical Center have developed FindAdapt, a Python package for fast and accurate detection of adapter patterns without relying on prior information. The researchers demonstrated that FindAdapt was far superior to existing approaches. It identified adapters successfully in 180 simulation datasets with diverse read structures and 3,184 real datasets covering a variety of commercial and customized small RNA library preparation kits. FindAdapt is stand-alone software that can be easily integrated into small RNA sequencing analysis pipelines.
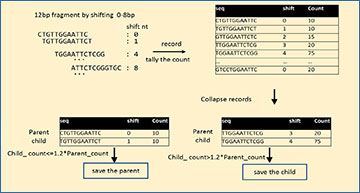
An overview of FindAdapt workflow
Availability – FindAdapt is freely available at https://github.com/chc-code/FindAdapt.
Chen H-C, Wang J, Shyr Y, Liu Q (2024) FindAdapt: A python package for fast and accurate adapter detection in small RNA sequencing. PLoS Comput Biol 20(1): e1011786. [article]