In genomics, understanding how genes are expressed in different cell types within their spatial context is a fundamental goal. With the advent of SPADE (SPAtial DEconvolution), a revolutionary method, researchers at the University of Arizona now have a powerful tool to unravel these spatial patterns and gain deeper insights into complex biological systems.
SPADE takes a multi-faceted approach by integrating spatial information into the analysis of cell type composition. Leveraging single-cell RNA sequencing, spatial transcriptomics, and histological data, SPADE accurately estimates the proportions of different cell types in various locations within tissues.
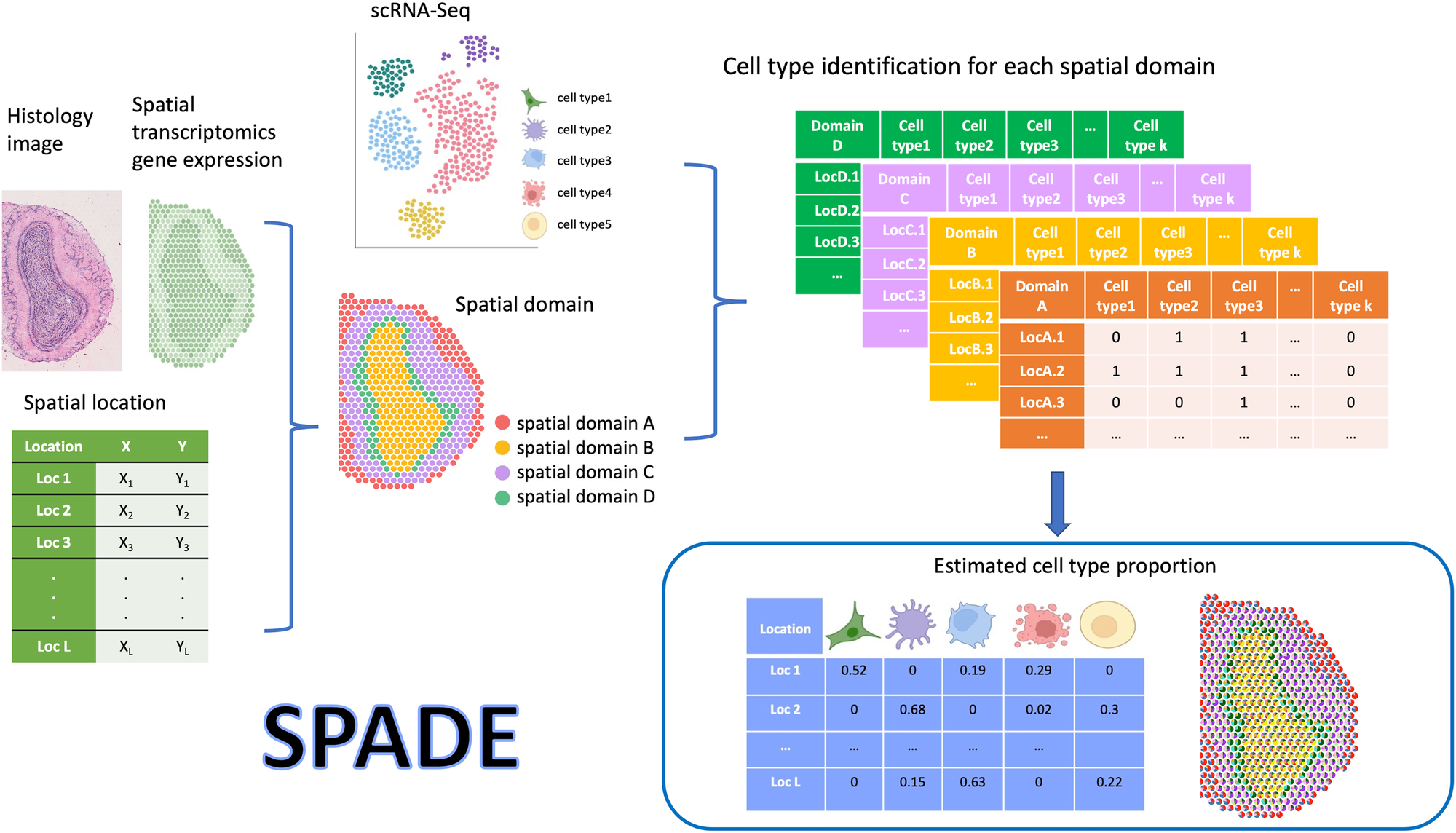
Schematic overview of SPADE
SPADE leverages reference single-cell RNA sequencing data to determine the cell type proportion at each location in the sample. To achieve this, SPADE first uses a combination of histology, spatial location, and gene expression information to identify spatial domains within a tissue. Subsequently, it performs a cell type selection for each domain by identifying the specific cell types present. Once the cell type information is obtained, SPADE utilizes scRNA-seq data to perform deconvolution, resulting in the estimation of cell type proportions for every spatial location. The final outcome of SPADE is the calculated cell type proportions for every spatial location.
In synthetic data simulations, SPADE has demonstrated its remarkable ability to discern cell type-specific spatial patterns effectively. But its true potential shines when applied to real-life datasets, where it offers invaluable insights into the dynamics of cellular interactions within tumor tissues.
By providing a deeper understanding of cellular diversity and dynamics, SPADE opens new avenues for exploring the complexities of biological systems. Researchers can now explore deeper into the interplay between different cell types and their spatial organization within tissues.
Moreover, SPADE represents a significant advancement in deciphering spatial gene expression patterns. By shedding light on the spatial distribution of gene expression, SPADE enables researchers to uncover previously hidden insights into the molecular mechanisms underlying various biological processes.
Availability – The R package and tutorial for implement SPADE is freely available on GitHub (https://github.com/anlingUA/SPADE).
Lu Y, Chen QM, An L. (2024) SPADE: spatial deconvolution for domain specific cell-type estimation. Commun Biol 7(1):469. [article]