When next generation sequencing is performed in large batches, there are several stages at which samples can be swapped or mislabeled. It is therefore helpful, when possible, to integrate measures into analysis pipelines to confirm that samples match their assigned metadata. Researchers from the Jackson Laboratory for Mammalian Genetics have developed RNA Strain-Match, a quality control tool developed to match RNA data in the form of sequence alignment files (i.e. SAM or BAM files) to their corresponding genotype without the use of an RNA variant call format file. The researchers successfully used RNA Strain-Match in tandem with assessment of markers for sex and transgene status to identify and correct sample mismatches in 50/379 samples (13%) from two distinct recombinant inbred mouse models (BXD and Collaborative Cross). They believe this tool will be beneficial to any research group working with similar data.
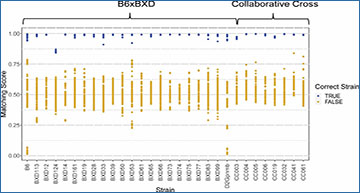
Matching scores by sample strain
Distributions of matching scores are plotted for each strain by correct sample strain (x-axis). The corresponding strain is shown in blue, while all other strains are shown in goldenrod.
Availability – https://github.com/jon-willcox/RNA-strain-match
Willcox JAL, Telpoukhovskaia MA, Hadad N, Boas SM, Dunn A, Saul MC, Ashbrook DG, Williams RW, O’Connell KMS, Kaczorowski CC. (2023) RNA Strain-Match: A tool for matching single-nucleus, single-cell, or bulk RNA-sequencing alignment data to its corresponding genotype. bioRXiv [online preprint]. [article]