Researchers at the Johns Hopkins University have developed ORFanage, a system designed to assign open reading frames (ORFs) to both known and novel gene transcripts while maximizing similarity to annotated proteins. The primary intended use of ORFanage is the identification of ORFs in the assembled results of RNA sequencing (RNA-seq) experiments, a capability that most transcriptome assembly methods do not have. These experiments demonstrate how ORFanage can be used to find novel protein variants in RNA-seq datasets, and to improve the annotations of ORFs in tens of thousands of transcript models in the RefSeq and GENCODE human annotation databases. Through its implementation of a highly accurate and efficient pseudo-alignment algorithm, ORFanage is substantially faster than other ORF annotation methods, enabling its application to very large datasets. When used to analyze transcriptome assemblies, ORFanage can aid in the separation of signal from transcriptional noise and the identification of likely functional transcript variants, ultimately advancing our understanding of biology and medicine.
The algorithm implemented in ORFanage
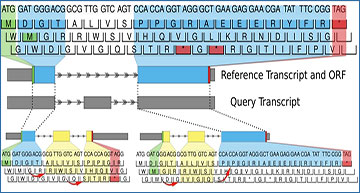
ORFanage begins by computing overlaps between reference ORF and query transcript. In the figure, dashed lines are used to connect matching intervals. For each overlap it extends coordinates towards the 3’ and 5’ ends based on suitable parameters. During extension, any changes to the exon structure may introduce shifting of the original frame (as indicated by red arrows). Once all intervals have been evaluated, ORFanage compares the results and reports the one with the highest score. In the figure, matching residues to the reference are highlighted in blue, and mismatching residues are highlighted in yellow. In this example, ORFanage selects the longer ORF on the lower right, which has 13 out of 17 matching residues, compared to the ORF on the lower left with only 4 out of 17 matching residues.
Availability – The code and test data are available for download on GitHub: https://github.com/alevar/ORFanage.
Varabyou A, Erdogdu B, Salzberg SL, Pertea M. (2023) Investigating Open Reading Frames in Known and Novel Transcripts using ORFanage. bioRXiv [online preprint]. [article]