Unraveling the mysteries of RNA has always been a challenging task, but recent advancements in technology have opened up new possibilities, allowing scientists to delve deeper into the secrets of RNA molecules.
Direct RNA sequencing is a cutting-edge technique that offers a unique advantage: the ability to simultaneously identify both the canonical bases and epitranscriptomic modifications present in each individual RNA molecule. This means that researchers can now explore not only the genetic code encoded in RNA but also the chemical modifications that adorn it.
However, despite the potential of direct RNA sequencing, progress has been hindered by the lack of biologically realistic training data. Without samples that accurately reflect the molecular landscape of RNA modifications, developing computational methods for analyzing this data has proven challenging.
In a groundbreaking study, researchers at the University of Heidelberg have addressed this limitation by synthesizing samples that mimic the complexity of natural RNA molecules, complete with modification labels at the molecular level. This innovative approach has paved the way for the development of a bespoke algorithm called mAFiA (m6A Finding Algorithm).
mAFiA is designed to accurately detect single m6A nucleotides, a common type of RNA modification, in both synthetic RNAs and natural mRNA on a single-read level. By analyzing individual RNA molecules, mAFiA uncovers distinct modification patterns that would otherwise remain hidden when examined at the ensemble level.
Training on synthetic RNA
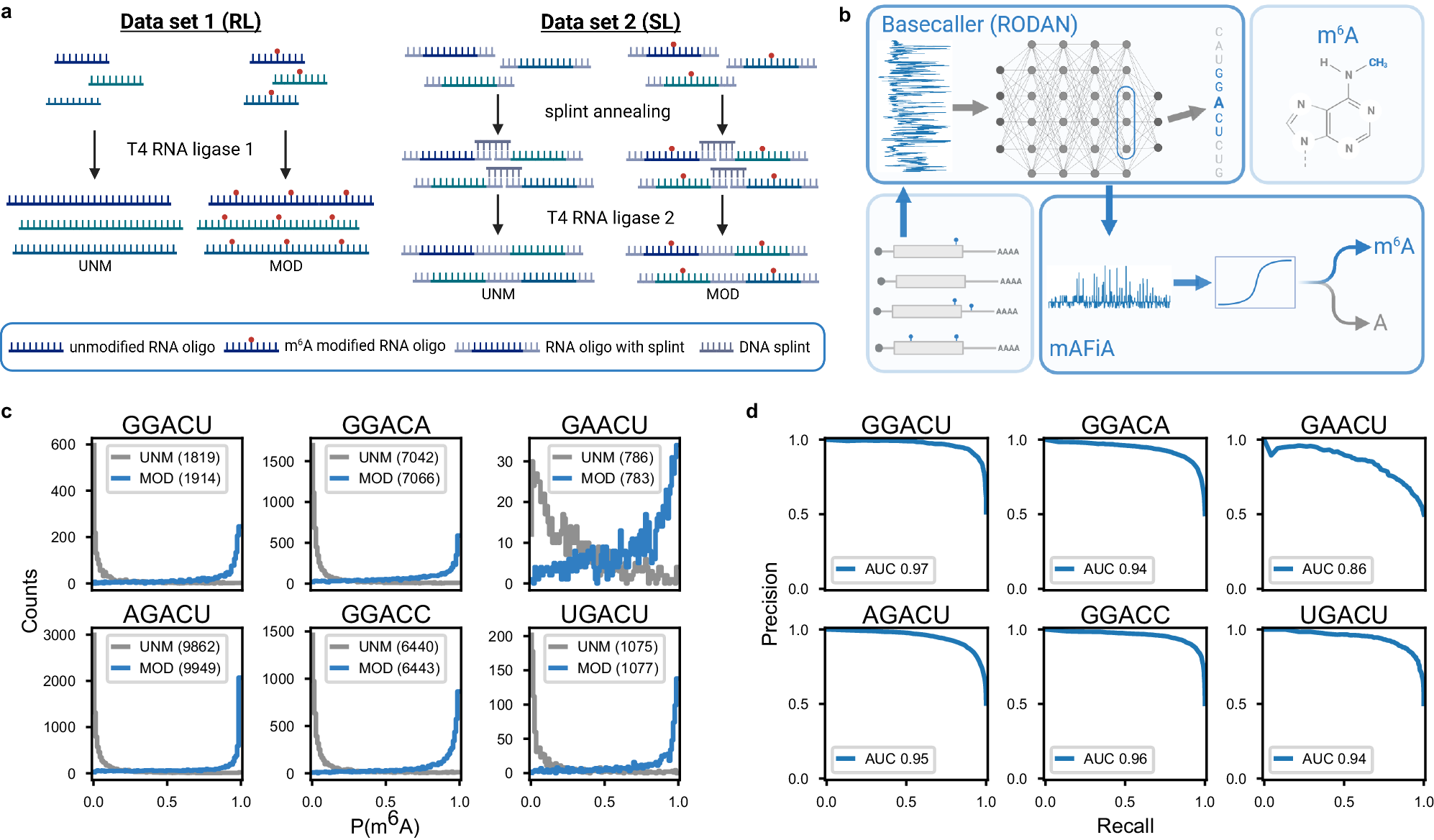
a Training data – Schematic representation of the two different ligation strategies – random ligation (RL) and splint ligation (SL). Left: 21 nt RNA oligos (colors indicate different sequences) with a central DRACH motif (A or m6A (red dot)) are concatenated to homopolymers by RNA ligase 1 and sequenced in unmodified (UNM) or modified (MOD) pools. Right: 33 nt RNA oligos with a central DRACH motif (A or m6A (red dot)) and flanking splint sequences (grey) are ligated to heteropolymers by splint-assisted ligation using RNA ligase 2 and sequenced in UNM or MOD pools. b Algorithm – From the backbone basecaller network RODAN7, mAFiA extracts a 768-dimensional feature vector ? that corresponds to a predicted nucleotide A in one of the target motifs. Logistic regression (see methods) is then applied to ? to generate the read-level m6A modification probability, ?(?6?), where 0≤?(?6?)≤1. c Model validation – mAFiA models trained on 75% of the RL dataset and validated on 25% of SL. The histogram shows the distribution of predicted ?(?6?) assigned to central A nucleotides mapped to various motifs, in unmodified (UNM) and modified (MOD) replicates. Gray lines correspond to nucleotides from the UNM sample and blue lines from MOD. Numbers in brackets are the validated sample size. d Precision-recall curves (PRCs) calculated from the ?(?6?) distributions in (c), with area-under-curve (AUC) given in the legend. Source data are provided as a Source Data file.
Moreover, compared to existing methods, mAFiA demonstrates superior accuracy in measuring site-level m6A stoichiometry in biological samples. This means that researchers can now obtain a more precise understanding of the abundance of m6A modifications at specific sites within RNA molecules.
By shedding light on the world of RNA modifications, scientists can gain deeper insights into the complex mechanisms that govern cellular processes. This knowledge not only enhances our understanding of basic biology but also holds promise for the development of novel therapeutic interventions targeting RNA-related diseases.
The synthesis of biologically realistic RNA samples and the development of advanced computational algorithms represent significant milestones in the field of RNA sequencing. With these tools at their disposal, researchers are poised to unlock new discoveries and push the boundaries of biological knowledge.
Availability -Software and models are available at: https://github.com/dieterich-lab/mAFiA.
Chan A, Naarmann-de Vries IS, Scheitl CPM, Höbartner C, Dieterich C. (2024) Detecting m6A at single-molecular resolution via direct RNA sequencing and realistic training data. Nat Commun 15(1):3323. [article]