Gene expression, the process by which information encoded in DNA is translated into functional molecules like proteins, is tightly regulated in all living organisms. While much attention is given to transcription, the process of making RNA from DNA, another crucial aspect of gene regulation occurs post-transcriptionally through mechanisms like RNA decay. RNA decay, the controlled breakdown of RNA molecules, plays a pivotal role in shaping gene expression in response to environmental stresses. In bacteria, RNA-binding proteins (RBPs) are key players in posttranscriptional regulation, yet their global impact on RNA stability remains poorly understood.
To investigate the influence of RBPs on RNA stability, researchers at the Helmholtz Centre for Infection Research conducted RNA sequencing experiments on Salmonella enterica bacteria over a time course following treatment with rifampicin, a drug that inhibits transcription initiation. This approach, known as RIF-seq, allowed them to measure changes in RNA stability in the presence and absence of the RBPs ProQ and CspC/E. By developing a sophisticated hierarchical Bayesian model, the researchers were able to correct for confounding factors and identify transcripts that decay differentially in response to the absence of these RBPs.
Pipeline and model description
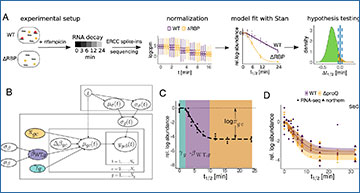
(A) RIF-seq workflow WT and ΔRBP strains are treated with rifampicin, cells are harvested at various time points and subjected to RNA-seq. Read counts are normalized before the extraction of biologically relevant parameters with a Bayesian model. Significant differences between strains are identified with Bayesian P values. (B) A plate diagram of the Bayesian models in this study. (C) Representative example of a decay curve in the LNM, illustrating regimes dominated by the different model parameters. The period of transcription elongation γ is marked in green, the exponential decay with decay rate β in purple and the constant regime governed by the fraction of baseline RNA in orange. (D) Comparison of RNA-seq and model fit with independent northern blot quantifications for SibC. (E) Hyperpriors and median of transcriptome-wide WT half-lives in the three Bayesian models. (F) Half-life distributions from the LNM for transcripts in selected COG categories. Adjusted p-values (compared to the first COG) were calculated using the Wilcoxon rank sum test.
The analysis revealed several key findings. Firstly, the researchers discovered that the median RNA half-life in Salmonella during early stationary phase is less than one minute, which challenges previous estimates and underscores the rapid turnover of RNA molecules in bacterial cells. Moreover, they found that over half of the 500 most long-lived transcripts are bound by at least one major RBP, suggesting a broad role for RBPs in shaping the bacterial transcriptome.
Further integration of differential stability estimates with CLIP-seq data, a technique that identifies RNA sequences bound by RBPs, uncovered that a significant portion of transcripts with ProQ and CspC/E binding sites decay differentially in the absence of these RBPs. Additionally, analysis of differentially destabilized transcripts revealed a specific role for ProQ in the oxidative stress response, highlighting the functional significance of RBPs in bacterial adaptation to environmental cues.
This study provides valuable insights into the intricate mechanisms of posttranscriptional gene regulation in bacteria. By elucidating the roles of RBPs such as ProQ and CspC/E in maintaining RNA stability, these researchers have deepened our understanding of how bacterial cells fine-tune gene expression in response to changing environmental conditions. Moving forward, further exploration of RNA decay pathways and RBP-mediated regulation promises to uncover new avenues for manipulating gene expression in bacterial pathogens and beyond.
Availability – Transcript annotations and source code for the Stan models have been made available at https://github.com/BarquistLab/RIF-seq_repo
Jenniches L, Michaux C, Popella L, Reichardt S, Vogel J, Westermann AJ, Barquist L. (2024) Improved RNA stability estimation through Bayesian modeling reveals most Salmonella transcripts have subminute half-lives. PNAS 121(14):e2308814121. [article]