Even for essential splice-site variants that are almost guaranteed to alter mRNA splicing, no current method can reliably predict whether exon-skipping, cryptic activation or multiple events will result, greatly complicating clinical interpretation of pathogenicity. Strikingly, ranking the four most common unannotated splicing events across 335,663 reference RNA-sequencing (RNA-seq) samples (300K-RNA Top-4) predicts the nature of variant-associated mis-splicing with 92% sensitivity. The 300K-RNA Top-4 events correctly identify 96% of exon-skipping events and 86% of cryptic splice sites for 140 clinical cases subject to RNA testing, showing higher sensitivity and positive predictive value than SpliceAI. Notably, RNA re-analyses showed missed 300K-RNA Top-4 events for several clinical cases tested before the development of this empirical predictive method. Simply, mis-splicing events that happen around a splice site in RNA-seq data are those most likely to be activated by a splice-site variant. The SpliceVault web portal allows users easy access to 300K-RNA for informed splice-site variant interpretation and classification.
Unannotated splicing events seen in 300K-RNA
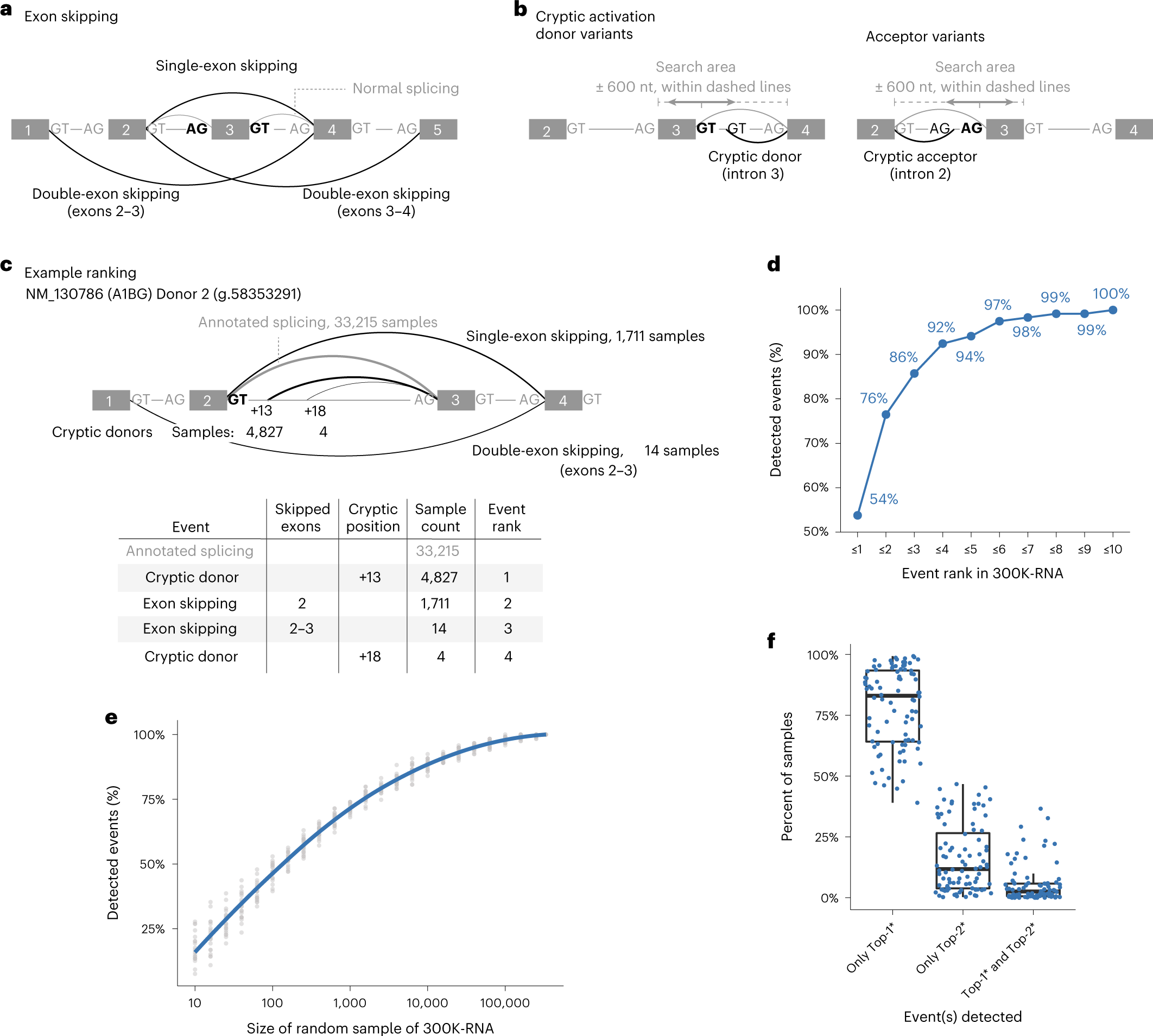
a, Exon-skipping events are evidenced by split-reads spanning nonconsecutive exons within the transcript. Splice sites (GT/AG motifs) shown in bold and black are those for which events are being ranked. b, Cryptic activation events are evidenced by split-reads spanning: (i) an annotated acceptor and an unannotated donor or (ii) an annotated donor and an unannotated acceptor. c, Example showing the Top-4* events for NM_130786 (A1BG) exon 2 donor (g.58353291). Exon/intron lengths are not drawn to scale. Arc thickness corresponds to event rank. d, One hundred percent (119/119) of exon-skipping and cryptic activation events detected across 88 variants are present in 300K-RNA, and 92% are in the Top-4* events for their respective splice site. e, Percent of the 119 true-positive events detected within random subsets of the 335,663 source specimens in 300K-RNA. Gray dots show proportion across 20 random samples; blue line shows mean proportions with LOESS smoothing. f, Top-1* and Top-2* events around the splice sites affected by our 88 variants typically occur in mutually exclusive specimens—with both events seen, on average, in only 5% of total samples where either event was detected. Internal lines of boxplot denote the median value, and the lower and upper limits of the boxes represent 25th and 75th percentiles. Whiskers extend to the largest and smallest values at most 1.5IQR. An asterisk indicates our filter for events involving skipping one or two exons and cryptic activation within 600 nt of the annotated splice site.
Availability – https://kidsneuro.shinyapps.io/splicevault/
Dawes R, Bournazos AM, Bryen SJ, Bommireddipalli S, Marchant RG, Joshi H, Cooper ST. (2023) SpliceVault predicts the precise nature of variant-associated mis-splicing. Nat Genet [Epub ahead of print]. [article]