Next generation RNA sequencing allows alternative splicing (AS) quantification with unprecedented resolution, with the relative inclusion of an alternative sequence in transcripts being commonly quantified by the proportion of reads supporting it as percent spliced-in (PSI). However, PSI values do not incorporate information about precision, proportional to the respective AS events’ read coverage. Beta distributions are suitable to quantify inclusion levels of alternative sequences, using reads supporting their inclusion and exclusion as surrogates for the two distribution shape parameters. Each such beta distribution has the PSI as its mean value and is narrower when the read coverage is higher, facilitating the interpretability of its precision when plotted.
Researchers at the Universidade de Lisboa have developed a computational pipeline, based on beta distributions accurately modelling PSI values and their precision, to quantitatively and visually compare AS between groups of samples. Thier methodology includes a differential splicing significance metric that compromises the magnitude of inter-group differences, the estimation uncertainty in individual samples, and the intra-group variability, being therefore suitable to multiple-group comparisons. To make their approach accessible and clear to both non-computational and computational biologists, the researchers developed betAS, an interactive web app and user-friendly R package for visual and intuitive differential splicing analysis from read count data.
Beta distributions model PSI levels and associated confidence 
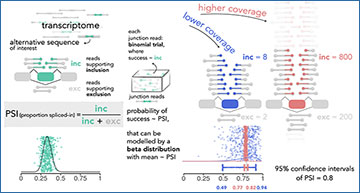
Explanatory diagram of the estimation of the proportion spliced-in (PSI) with the beta distribution for an alternative transcript sequence (exon) of interest (green) from the RNA-seq junction read counts supporting the sequence’s inclusion (inc) or exclusion (exc). The ability of the beta distribution to incorporate the mean value and the confidence of the PSI is further illustrated for two discrepant scenarios of read coverage: lower, with 10 reads (blue), and higher, with 1000 reads (salmon). PSIs randomly generated from the respective beta distributions (500 coloured vertically “jittered” points per distribution and associated solid density lines) are dispersed around mean values, being less dispersed as coverage (i.e., confidence associated to supporting evidence) increases. Coloured 95% confidence intervals for a proportion’s test with p (in this case, PSI) equal to 0.8, with 10 (blue) or 1000 (salmon) trials. Coloured (green/blue/salmon) squircles: alternative exons; grey squircles: constitutive exons; junction read depictions coloured according to their exon coverage.
Availability – betAS web app is publicly available at https://compbio.imm.medicina.ulisboa.pt/app/betAS. Its source code is available at https://github.com/DiseaseTranscriptomicsLab/betAS.
Ascensao-Ferreira M, Martins-Silva R, Saraiva-Agostinho N, Barbosa-Morais NL. (2024) betAS: intuitive analysis and visualisation of differential alternative splicing using beta distributions. RNA [Epub ahead of print]. [abstract]