Bacteria colonize almost all parts of the human body and can differ significantly. However, the population level transcriptomics measurements can only describe the average bacteria population behaviors, ignoring the heterogeneity among bacteria. Researchers from the Zhejiang University School of Medicine have developed a droplet-based high-throughput single-microbe RNA-seq assay (smRandom-seq), using random primers for in situ cDNA generation, droplets for single-microbe barcoding, and CRISPR-based rRNA depletion for mRNA enrichment. smRandom-seq showed a high species specificity (99%), a minor doublet rate (1.6%), a reduced rRNA percentage (32%), and a sensitive gene detection (a median of ~1000 genes per single E. coli). Furthermore, smRandom-seq successfully captured transcriptome changes of thousands of individual E. coli and discovered a few antibiotic resistant subpopulations displaying distinct gene expression patterns of SOS response and metabolic pathways in E. coli population upon antibiotic stress. smRandom-seq provides a high-throughput single-microbe transcriptome profiling tool that will facilitate future discoveries in microbial resistance, persistence, microbe-host interaction, and microbiome research.
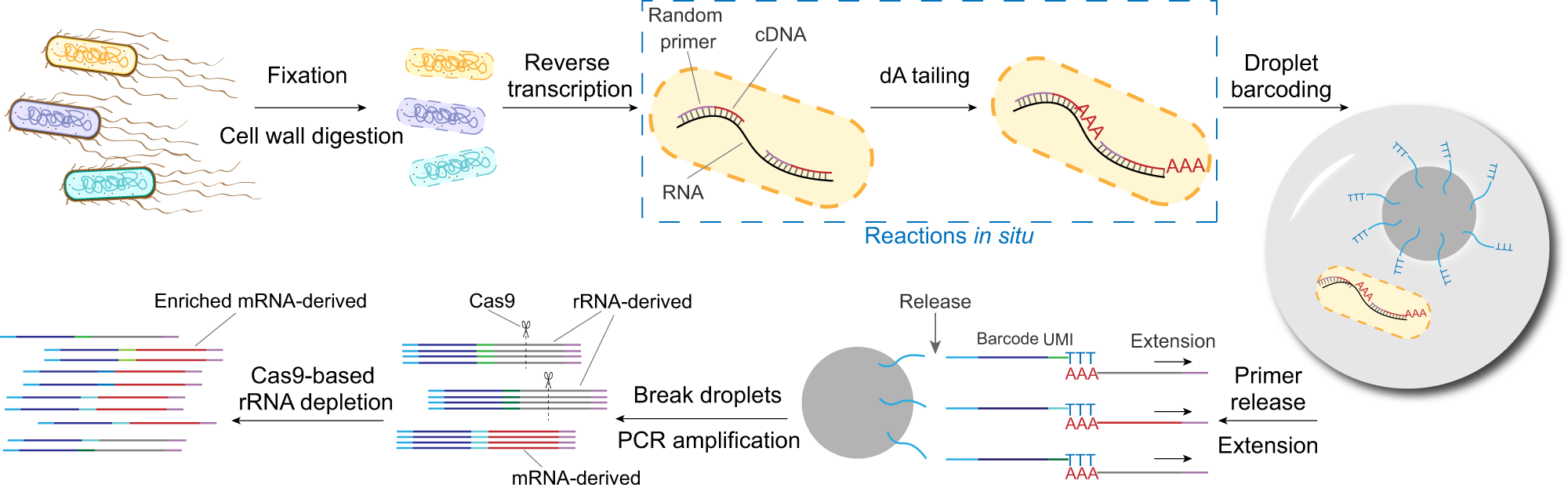
Overview of the droplet-based smRandom-seq
The workflow of smRandom-seq for microbe samples includes fixation, cell wall digestion, reverse transcription, dA tailing, droplet barcoding, primers releasing and extension, droplets breaking and PCR amplification, and Cas9-based rRNA depletion and sequencing. Blue dashed box: the two in situ reactions, including reverse transcription and dA tailing. AAA: dA tail in the 3’ of cDNA, TTT: poly(dT) in the barcoded primers.
Xu Z, Wang Y, Sheng K, Rosenthal R, Liu N, Hua X, Zhang T, Chen J, Song M, Lv Y, Zhang S, Huang Y, Wang Z, Cao T, Shen Y, Jiang Y, Yu Y, Chen Y, Guo G, Yin P, Weitz DA, Wang Y. (2023) Droplet-based high-throughput single microbe RNA sequencing by smRandom-seq. Nat Commun 14(1):5130. [article].