UT Southwestern-led study produces new tool for cancer-fighting precision medicine
UT Southwestern researchers aiming to better understand how cancer cells interact with immune cells used a method called single cell RNA sequencing. (Photo credit: Getty Images)
“Immunotherapies have made incredible strides in extending survival for cancer patients, but they only work about 20% of the time. To make immunotherapies more beneficial, we need to have a much better understanding of the cellular composition of specific tumors and how those cells interact with each other,” said Isaac Chan, M.D., Ph.D., Assistant Professor of Internal Medicine and Molecular Biology and in the Harold C. Simmons Comprehensive Cancer Center at UT Southwestern.
Dr. Chan explained that over the past several decades, researchers have had a growing understanding that malignant tumors aren’t just made of cancer cells but have a variety of cell types, including immune cells that can either hinder or help tumors thrive. Existing immunotherapies take advantage of this phenomenon by stimulating T cells present in and around tumors to attack cancer cells.
However, T cells aren’t the only cancer-fighting cells present in tumors that change after interacting with cancer cells, Dr. Chan noted. Natural killer (NK) cells, part of the innate immune system, can also help fight cancer. But the Chan Lab and others have shown that interactions between cancer cells and NK cells can exhaust or even reprogram – or corrupt – NK cells to promote tumors instead of killing them. Dr. Chan and his team were able to identify these reprogrammed NK cells in breast tumors in previous research. Patients who had high levels of reprogrammed NK cells had worse outcomes, suggesting the potential for therapeutically targeting these cells.
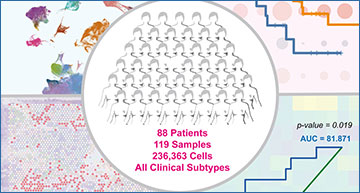
To better understand how cancer cells interact with immune cells, Dr. Chan and his colleagues took advantage of a method called single cell RNA sequencing (scRNA-seq), which evaluates gene expression in each individual cell. By combining several publicly available scRNA-seq databases, they created a new dataset that included 119 tumor samples from 88 breast cancer patients. Using this larger database, the researchers were able to analyze data from 236,363 cells, far more than any breast cancer scRNA-seq analysis thus far.
Using a computational technique, the team grouped together cancer cells in which similar sets of genes were turned on or off and found there were actually 10 categories of breast cancer cells. Just three categories of breast cancer cells (hormone positive, HER2 positive, and triple negative) are typically used to determine appropriate therapy. The distribution of the 10 categories of cancer cells varied among tumors; nearly all tumors had a mix of different subtypes, Dr. Chan said.
Dr. Chan noted that this approach – evaluating interactions between cancer and immune cells through their gene expression – could assess the likelihood of response to different immunotherapies across a wide variety of cancer types, helping doctors choose patients for whom this treatment will be the most successful.
Source – UT Southwestern
Availability – https://github.com/ChanLab-UTSW/BreastCancer_Integrated
Xu L, Saunders K, Huang SP, Knutsdottir H, Martinez-Algarin K, Terrazas I, Chen K, McArthur HM, Maués J, Hodgdon C, Reddy SM, Roussos Torres ET, Xu L, Chan IS. (2024) A comprehensive single-cell breast tumor atlas defines epithelial and immune heterogeneity and interactions predicting anti-PD-1 therapy response. Cell Rep Med [Epub ahead of print]. [article]