Emerging single-cell technologies provide high-resolution measurements of distinct cellular modalities opening new avenues for generating detailed cellular atlases of many and diverse tissues. The high dimensionality, sparsity, and inaccuracy of single cell sequencing measurements, however, can obscure discriminatory information, mask cellular subtype variations and complicate downstream analyses which can limit our understanding of cell function and tissue heterogeneity.
Researchers at the University of New South Wales have developed a novel pre-processing method (scPSD) inspired by power spectral density analysis that enhances the accuracy for cell subtype separation from large-scale single-cell omics data. The researchers comprehensively benchmarked their method on a wide range of single-cell RNA-sequencing datasets and showed that scPSD pre-processing, while being fast and scalable, significantly reduces data complexity, enhances cell-type separation, and enables rare cell identification. Additionally, they applied scPSD to transcriptomics and chromatin accessibility cell atlases and demonstrated its capacity to discriminate over 100 cell types across the whole organism and across different modalities of single-cell omics data.
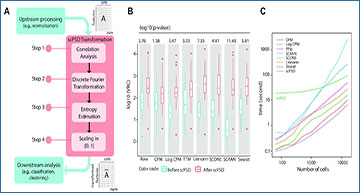
Overview of scPSD and performance evaluation on scRNA-seq datasets
(A) the scPSD transformation framework comprising four consecutive steps of feature extraction and standardization. scPSD can fit into a single-cell sequencing analysis pipeline after the upstream processing (or directly on raw data) to enhance downstream analyses. (B) box plots comparing VRC (variance ratio criterion) as a measure of how well-formed distinct cell-types are before/after scPSD transformation of normalized and raw counts across 25 curated scRNA-seq datasets. (C) computational runtime of scPSD and normalization methods as scales with increasing number of cells. (D) heatmaps representing accuracy of cell-type prediction—for each of 25 scRNA-seq datasets—on 20% randomly held out data (test set) after training SVM (support vector machine) and KNN (k-nearest neighbor) models on remaining 80% of data (training set), before and after scPSD transformation. (E) SVM training time in second before and after scPSD transformation demonstrating significant reduction in convergence time after transformation. (F) heatmaps representing SVM test accuracy identifying rare cell-type identification—defined as the smallest cell-type population constituting <1% to 14% of captured cells across 25 scRNA-seq datasets—before and after scPSD transformation. (G) SVM test accuracy upon increasing feature coverage using ‘deng reads’ dataset.
Availability – The scPSD method has been implemented in MATLAB (https://github.com/VafaeeLab/psdMAT), Python (https://github.com/VafaeeLab/psdPy) and R package (https://github.com/VafaeeLab/psdR).
Zandavi SM, Koch FC, Vijayan A, Zanini F, Mora FV, Ortega DG, Vafaee F. (2022) Disentangling single-cell omics representation with a power spectral density-based feature extraction. Nucleic Acids Research [Epub ahead of print]. [article]