Cancer is not a single disease but a diverse group of diseases characterized by distinct molecular profiles. Accurately classifying cancer subgroups is crucial for developing personalized treatment strategies tailored to individual patients. Recent advancements in high-throughput sequencing technologies have allowed researchers to generate vast amounts of transcriptomic data from cancer samples. Leveraging this wealth of information, computational methods are being developed to improve cancer subtyping and enhance personalized medicine approaches.
Understanding Meningioma Classification
In a recent study, researchers from the Baylor College of Medicine focused on meningioma, a type of brain tumor, to evaluate different feature selection schemes for cancer classification. Meningiomas exhibit heterogeneity, with various molecular subtypes, including benign and malignant classes. The study aimed to develop a computational algorithm that could accurately classify meningioma subgroups based on transcriptomic data.
Evaluation of Feature Selection Schemes
The researchers evaluated different feature selection schemes using bulk transcriptomic data from 78 meningioma samples. While some schemes showed good classification accuracy, they also exhibited confusion between malignant and benign molecular classes in approximately 8% of the samples. To address this challenge, the study explored the use of single-cell transcriptomic data (~10K cells) to improve subgroup resolution.
Development of CLIPPR Algorithm
To integrate interpretable features from both bulk and single-cell profiling, the researchers developed an algorithm called CLIPPR. CLIPPR combines top-performing single-cell models with RNA-inferred copy number variation (CNV) signals and the initial bulk model to create a meta-model. This meta-model exhibited the strongest performance in meningioma classification, surpassing the accuracy of individual models.
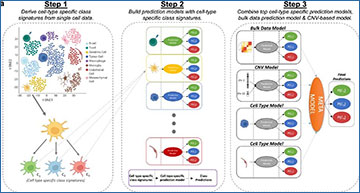
Overview of the CLIPPR algorithm
Step 1 involves deriving cell type-specific class signatures from single-cell meningioma data. In Step 2, prediction models are constructed using cell type-specific class features (ctRFs). For each feature set, the bulk RNA-Seq data was used to train a cell-type specific Random Forest model. Models were also constructed using RNA-inferred CNV signals (cnvRF) and the bulk model (bulkRF). In Step 3, a meta-model is assembled, integrating the top cell type-specific models, the bulk model, and the CNV model.
Validation and Generalizability
The performance of CLIPPR was validated on a larger dataset consisting of 792 bulk meningioma samples gathered from multiple institutions. The algorithm demonstrated superior overall accuracy and effectively resolved confusion between benign and malignant subgroups. Additionally, the generalizability of CLIPPR was assessed using single-cell and bulk transcriptomic data from glioma samples, further confirming its efficacy in cancer subtyping.
The development of computational algorithms like CLIPPR represents a significant advancement in cancer subtyping. By integrating single-cell and bulk transcriptomic data, CLIPPR enables improved classification of cancer subgroups, providing valuable insights into their biology. Ultimately, algorithms like CLIPPR have the potential to enhance personalized medicine approaches by guiding more precise and effective treatment strategies for cancer patients.
Shetty A, Wang S, Khan AB, English CW, Nouri SH, Magill ST, Raleigh DR, Klisch TJ, Harmanci AO, Patel AJ, Harmanci AS. (2024) Leveraging Single-Cell Sequencing to Classify and Characterize Tumor Subgroups in Bulk RNA-Sequencing Data. bioRXiv [Epub ahead of print]. [article]