In the fields of biology and genetics, single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool for understanding the complex landscape of cellular diversity within tissues and organs. However, annotating the multitude of cell types identified through scRNA-seq analysis is a laborious and time-consuming task. Enter GPT-4, the latest large language model, which promises to revolutionize cell type annotation by accurately leveraging marker gene information. Researchers at Columbia University and the Duke University School of Medicine demonstrate that GPT-4, the fourth iteration of the Generative Pre-trained Transformer series, has remarkable proficiency in annotating cell types using marker gene information derived from scRNA-seq data. Across hundreds of tissue and cell types, GPT-4 consistently generates cell type annotations that exhibit strong concordance with manual annotations performed by expert biologists. This breakthrough has the potential to significantly reduce the effort and expertise required for cell type annotation, making scRNA-seq analysis more accessible to researchers across diverse fields.
The key innovation lies in GPT-4’s ability to process marker gene information and accurately assign cell type labels to individual cells within scRNA-seq datasets. By leveraging its vast knowledge base and pattern recognition capabilities, GPT-4 can swiftly and accurately categorize cells into their respective types, facilitating deeper insights into cellular composition and function.
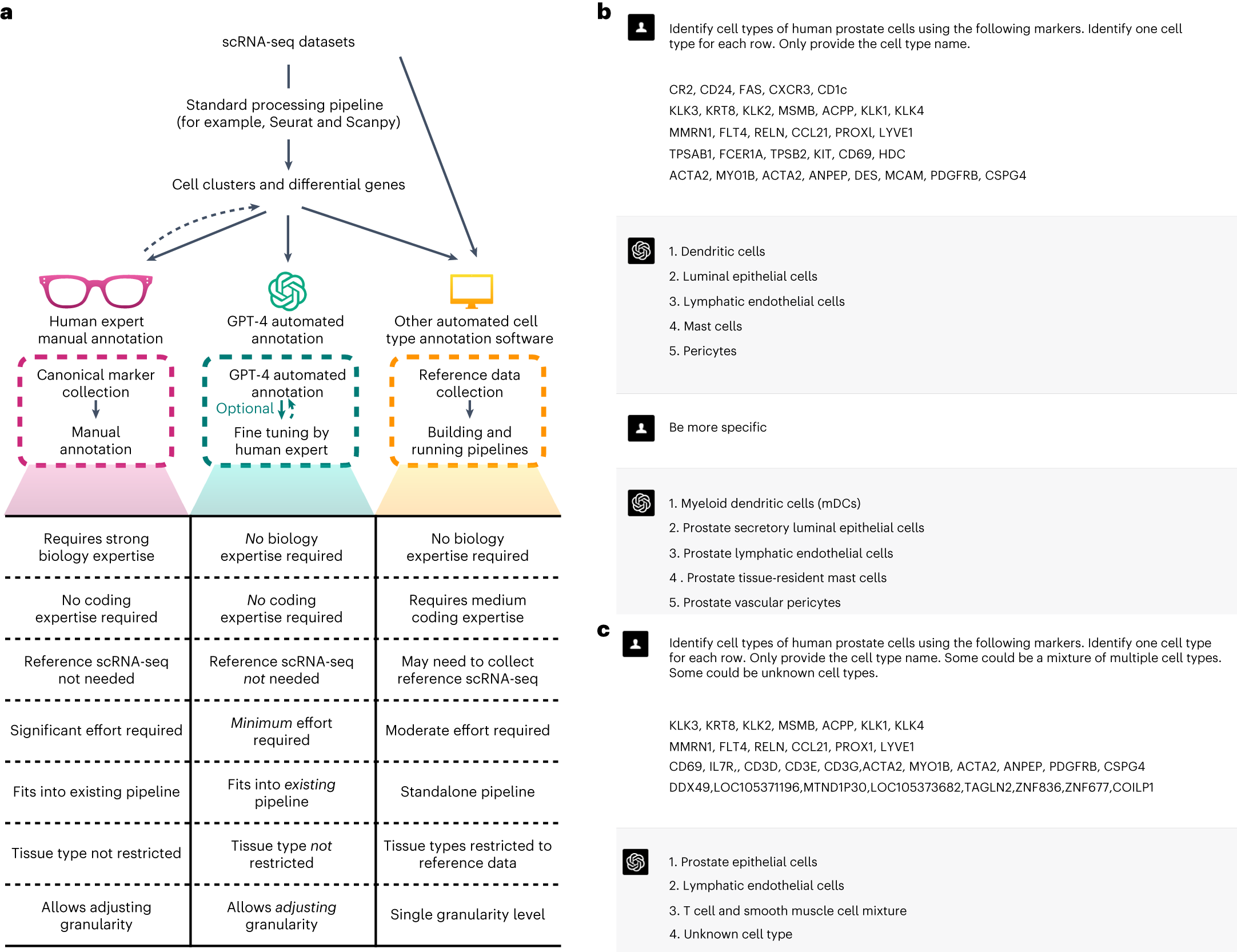
To further democratize access to GPT-4’s cell type annotation capabilities, the researchers have developed an R software package called GPTCelltype. This user-friendly tool harnesses the power of GPT-4 for automated cell type annotation, allowing researchers to seamlessly integrate this cutting-edge technology into their scRNA-seq analysis pipelines. With GPTCelltype, even researchers with limited computational expertise can harness the full potential of GPT-4 for cell type annotation, accelerating the pace of discovery in biological research.
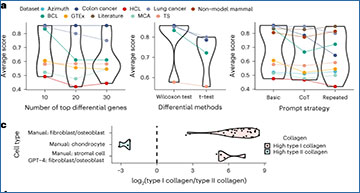
Examples of GPT-4’s cell type annotation and comparisons with other methods
a, Comparison of cell type annotations by human experts, GPT-4, and other automated methods. b, Example of GPT-4 annotating human prostate cells with increasing granularity. c, Example of GPT-4 annotating single, mixed and new cell types
The advent of GPT-4 and tools like GPTCelltype holds profound implications for the field of single-cell biology. By automating the cell type annotation process, researchers can expedite the analysis of scRNA-seq data and uncover novel insights into cellular heterogeneity and function. Moreover, GPT-4’s ability to accurately annotate cell types across diverse tissue types opens up new avenues for investigating complex biological phenomena and disease mechanisms.
GPT-4 represents a paradigm shift in the field of single-cell RNA sequencing analysis. By harnessing the power of natural language processing and machine learning, GPT-4 offers a transformative solution to the challenge of cell type annotation. With its ability to accurately categorize cells across diverse tissues and its user-friendly implementation through GPTCelltype, GPT-4 promises to accelerate discoveries in single-cell biology and pave the way for a deeper understanding of cellular dynamics in health and disease.
Availability – The GPTCelltype package (v.1.0.0) is provided as an open-source software package with a detailed user manual available in the GitHub repository at https://github.com/Winnie09/GPTCelltype. The software is released in Zenodo under https://doi.org/10.5281/zenodo.8317406 for all versions.
Hou W, Ji Z. (2024) Assessing GPT-4 for cell type annotation in single-cell RNA-seq analysis. Nat Methods [Epub ahead of print]. [article]