Annotating cell types within single-cell RNA sequencing (scRNA-seq) datasets has posed a significant challenge, especially as new data continuously emerges. To address this pressing issue, researchers at Peking University have developed a groundbreaking tool called CANAL, offering a novel approach to cell-type annotation that evolves with the ever-expanding landscape of scRNA-seq data.
Traditionally, cell-type annotation tools have relied on well-annotated datasets, lacking the ability to adapt to new information. CANAL revolutionizes this approach by leveraging transformer-based pre-trained language models, which have demonstrated remarkable capabilities in single-cell biology research. By continuously fine-tuning these models on a vast amount of unlabeled scRNA-seq data, CANAL ensures that it stays abreast of the latest advancements in cell-type identification.
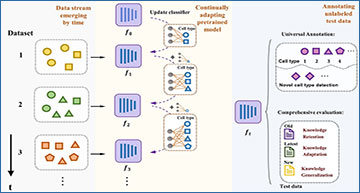
Overview of CANAL
The well-annotated data stream evolves over time, and the PTLM undergoes continuous adaptation at different training stages. It is not necessary for the cell-type set to be fully overlapped at each stage. When new cell types emerge in a new stage, our classifier is updated to encompass the entire set of encountered cell types. The fine-tuned model, denoted as ft, exhibits sufficient generalizability to directly annotate diverse unlabeled test datasets, thereby achieving universal annotation.
One of the key features of CANAL is its ability to mitigate catastrophic forgetting, a phenomenon where previous knowledge is lost when new information is learned. To address this, CANAL employs an experience replay schema, which continuously reviews crucial examples from previous training stages. This dynamic example bank ensures that important cell-type-specific information is retained, even as the model evolves.
Furthermore, CANAL utilizes representation knowledge distillation to maintain consistency between previous and current models, preserving valuable insights gleaned from past training stages. This ensures that knowledge learned throughout the continuous fine-tuning process is retained and not lost over time.
Additionally, CANAL’s universal annotation framework allows for the seamless incorporation of new cell types as they emerge. By absorbing new cell types from newly arrived, well-annotated training datasets, as well as automatically identifying novel cells in unlabeled datasets, CANAL ensures that its cell-type annotation library remains comprehensive and up-to-date.
Comprehensive experiments conducted with CANAL have demonstrated its versatility and high model interpretability across various biological scenarios. By combining the power of pre-trained language models with continuous learning, CANAL represents a significant advancement in the field of cell-type annotation, paving the way for deeper insights into the complex world of single-cell biology.
Availability – An implementation of CANAL is available from https://github.com/aster-ww/CANAL-torch.
Wan H, Yuan M, Fu Y, Deng M. (2024) Continually adapting pre-trained language model to universal annotation of single-cell RNA-seq data. Brief Bioinform 25(2):bbae047. [article]