Modifications on viral RNAs (vRNAs), either genomic RNAs or RNA transcripts, have complex effects on the viral life cycle and cellular responses to viral infection. The advent of Oxford Nanopore Technologies Direct RNA Sequencing provides a new strategy for studying RNA modifications. To this end, multiple computational tools have been developed, but a systemic evaluation of their performance in mapping vRNA modifications is lacking. Researchers at the City University of Hong Kong tested 10 computational tools using the Sindbis virus (SINV) RNAs isolated from infected mammalian (BHK-21) or mosquito (C6/36) cells, with in vitro-transcribed RNAs serving as modification-free control. Three single-mode approaches were shown to be inapplicable in the viral context, and three out of seven comparative methods required cutoff adjustments to reduce false-positive predictions. Utilizing optimized cutoffs, an integrated analysis of comparative tools suggested that the intersected predictions of Tombo_com and xPore were significantly enriched compared with the background. Consequently, a pipeline integrating Tombo_com and xPore was proposed for vRNA modification detection; the performance of which was supported by N6-methyladenosine prediction in severe acute respiratory syndrome coronavirus 2 RNAs using publicly available data. When applied to SINV RNAs, this pipeline revealed more intensive modifications in subgenomic RNAs than in genomic RNAs. Modified uridines were frequently identified, exhibiting substantive overlapping between vRNAs generated in different cell lines. On the other hand, the interpretation of other modifications remained unclear, underlining the limitations of the current computational tools despite their notable potential.
Proposal of a vRNA modification detection pipeline and its performance evaluation
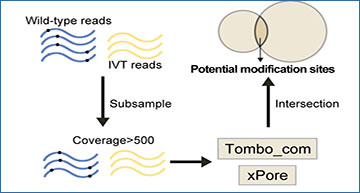
(A) Proposed pipeline for detecting vRNA modifications utilizing computational methods. Briefly, native and IVT reads are subsampled to an equal coverage (>500), followed by Tombo_com and xPore analyses and intersection identification. Cutoff values at the coverage depth of 500 are recommended for use. (B) Venn diagram showing the relationship between modification sites detected at different coverage depths while using the same cutoff values. Different numbers of reads (500, 1,000, and 1,500) were randomly subsampled from the IVT and BHK-21 full-length viral reads and analyzed the Tombo_com and xPore pipeline. (C) Detection of modification sites in the SARS-CoV-2 vRNAs using the Tombo_com and xPore pipeline. (D) Comparison between the computational pipeline outputs and MeRIP data in detecting SARS-CoV-2 vRNA m6A modifications. Modified A sites were extracted from the total prediction result of the Tombo_com and xPore pipeline. Caco-2 and Vero indicate MeRIP data measured in Caco-2 and Vero cells, respectively.
Tan L, Guo Z, Wang X, Kim DY, Li R. (2024) Utilization of nanopore direct RNA sequencing to analyze viral RNA modifications. mSystems [Epub ahead of print]. [article]