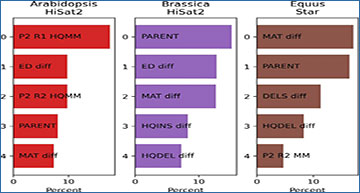
Understanding how genes are expressed and regulated is essential for unraveling the mysteries of life. One fascinating phenomenon that researchers have been exploring is parent-of-origin allele-specific gene expression (ASE) in interspecies hybrids. This refers to the differential expression of genes depending on whether they are inherited from the mother or the father.
A recent study by researchers at West Virginia University delved into this phenomenon using RNA sequencing (RNA-seq) data from interspecies hybrids. RNA-seq is a powerful tool that allows scientists to measure gene expression by analyzing the transcripts present in a cell. In hybrids, where genetic material from different species is mixed, detecting ASE can provide insights into how genes are regulated and expressed.
However, analyzing RNA-seq data from hybrids poses challenges, particularly when it comes to aligning the reads to the correct parental reference genomes. Traditional alignment methods may struggle to accurately assign reads to their proper references, leading to inaccuracies in ASE detection.
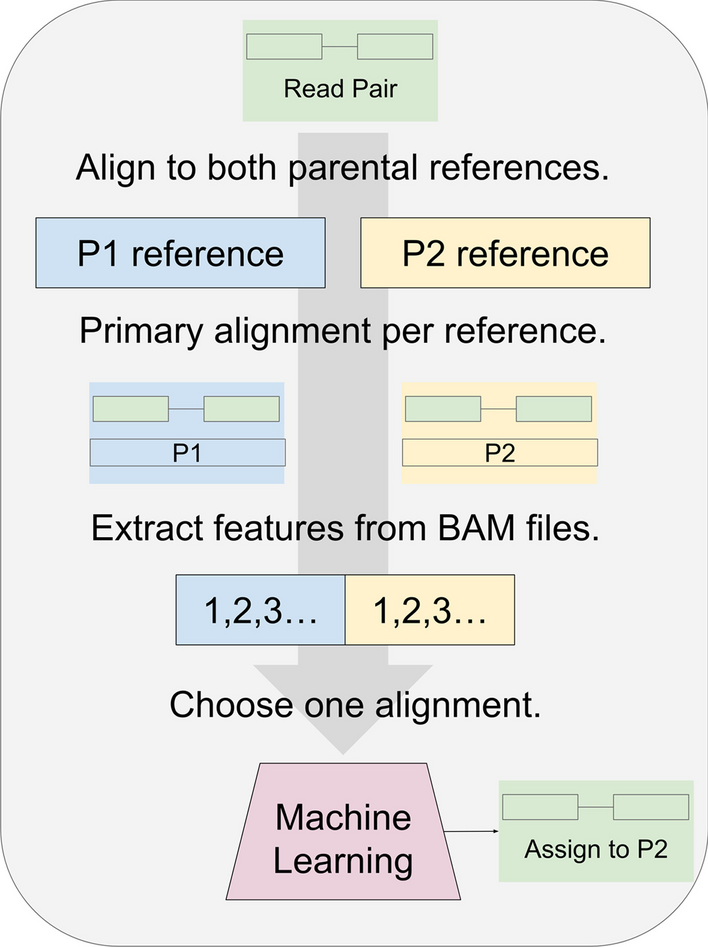
To address this issue, the researchers turned to machine learning techniques. They developed a novel approach that combines alignment-based methods with a machine learning post-processor. This post-processor extracts alignment features from the data and trains a random forest classifier to determine the correct parent of origin for each RNA-seq read pair.
Process for classifying RNA-seq by machine learning
For training the model, read pairs are obtained from parents P1 and P2. Each read pair is aligned to the P1 and P2 references separately. A filter removes all but the primary alignment per read pair. The process can use transcriptomes or genomes as references, and any suitable aligner. For read pairs that align to both P1 and P2, features are extracted and given to the machine learning model. Initially, the model is given the true parent label per read pair and trained to predict this. After training, read pairs from the hybrid cross are given instead. In this illustration, after a hybrid read pair (green) is aligned to the P1 (blue) and P2 (yellow) references, the trained model (red) chooses P2 as the more likely source. For most of this study, the model was a random forest binary classifier. Read pairs classified by this process could be binned by gene and quantified to detect allele-specific gene expression in the hybrid
While this study represents a significant step forward in ASE detection in interspecies hybrids, the researchers acknowledge that challenges may arise when applying this approach to real-world datasets. Nevertheless, their findings demonstrate the potential of machine learning to improve our ability to unravel complex genetic processes.
This research highlights the power of combining traditional bioinformatics methods with machine learning algorithms to tackle challenging biological questions. By harnessing the capabilities of artificial intelligence, scientists can unlock new insights into gene expression and regulation, paving the way for exciting discoveries in genetics and beyond.
Availability – See https://zenodo.org/records/10183055 for source code and notebooks.
Miller JR, Adjeroh DA. (2024) Machine learning on alignment features for parent-of-origin classification of simulated hybrid RNA-seq. BMC Bioinformatics 25(1):109. [article]