Cell-cell interactions (CCIs) and communication (CCC) play vital roles in orchestrating complex biological systems. Understanding how cells interact and communicate with each other is crucial for unraveling the underlying mechanisms of various physiological and pathological processes. With the advent of single-cell RNA sequencing (scRNA-seq) technology, researchers now have a powerful tool to delve deeper into these interactions by identifying ligand-receptor (LR) gene interactions between cells. However, existing methods for examining LR interactions often focus on individual pairs of genes, limiting their ability to capture the complexity of cell-cell interactions.
LR Hunting
Researchers at the University of Miami’s Miller School of Medicine have developed LR hunting, a novel computational approach designed to decipher cell-cell interactions by identifying significant LR interactions across different cell types simultaneously. The method utilizes a two-step process to analyze scRNA-seq data and unveil meaningful CCIs.
- Data Imputation with Random Forests (RFs): The first step involves data imputation using a random forests-based technique. This approach bridges the data between different cell types, ensuring robustness and accuracy in capturing LR interactions. By repeating the computation procedures multiple times and aggregating imputed minimal depth index (IMDI), LR hunting enhances the reliability of the imputation process.
- Identification of Significant LR Interactions: In the second step, LR hunting employs unsupervised RFs to identify significant LR interactions among all combinations of LR pairs simultaneously. This comprehensive analysis allows researchers to capture the complexity of cell-cell interactions and uncover biologically meaningful insights.
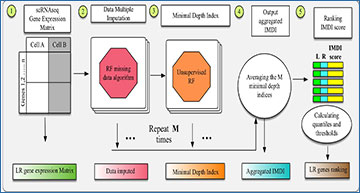
Illustration of RF methods
(A) Data sheet and imputation illustration. (B) The minimal depth of w in a maximal v-subtree. Letters in parent nodes identify the variable used to split the node. There are two maximal v-subtrees, marked in red. The maximal v-subtree on the left side is with terminal nodes 1 and 2; that on the right side is with terminal nodes 3, 4, 5, and 6. The minimal depth of w in the second maximal v-subtree is the depth of w (d = 2 marked with pink background) normalized by the subtree depth (m = 3), which is d/m = 2/3. (C) Model workflow for LR hunting.
Validation and Applications
LR hunting has been successfully validated using real-world datasets, including a mouse cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) dataset and a scRNA-seq dataset from triple-negative breast cancer. The results demonstrate the ability of LR hunting to recover biologically relevant CCIs and shed light on the intricate interplay between different cell types.
LR hunting represents a promising approach for deciphering cell-cell interactions using single-cell RNA sequencing data. By leveraging advanced computational techniques, LR hunting offers a comprehensive and robust method for identifying significant LR interactions across diverse cell populations. This innovative approach has the potential to advance our understanding of complex biological systems and pave the way for new discoveries in fields such as cancer biology, immunology, and developmental biology.
Availability – LR hunting analysis code is available is at https://github.com/TransBioInfoLab/LRinteractions.
Lu M, Sha Y, Silva TC, Colaprico A, Sun X, Ban Y, Wang L, Lehmann BD, Chen XS. (2024) LR Hunting: A Random Forest Based Cell-Cell Interaction Discovery Method for Single-Cell Gene Expression Data. Front Genet 12:708835. [article]