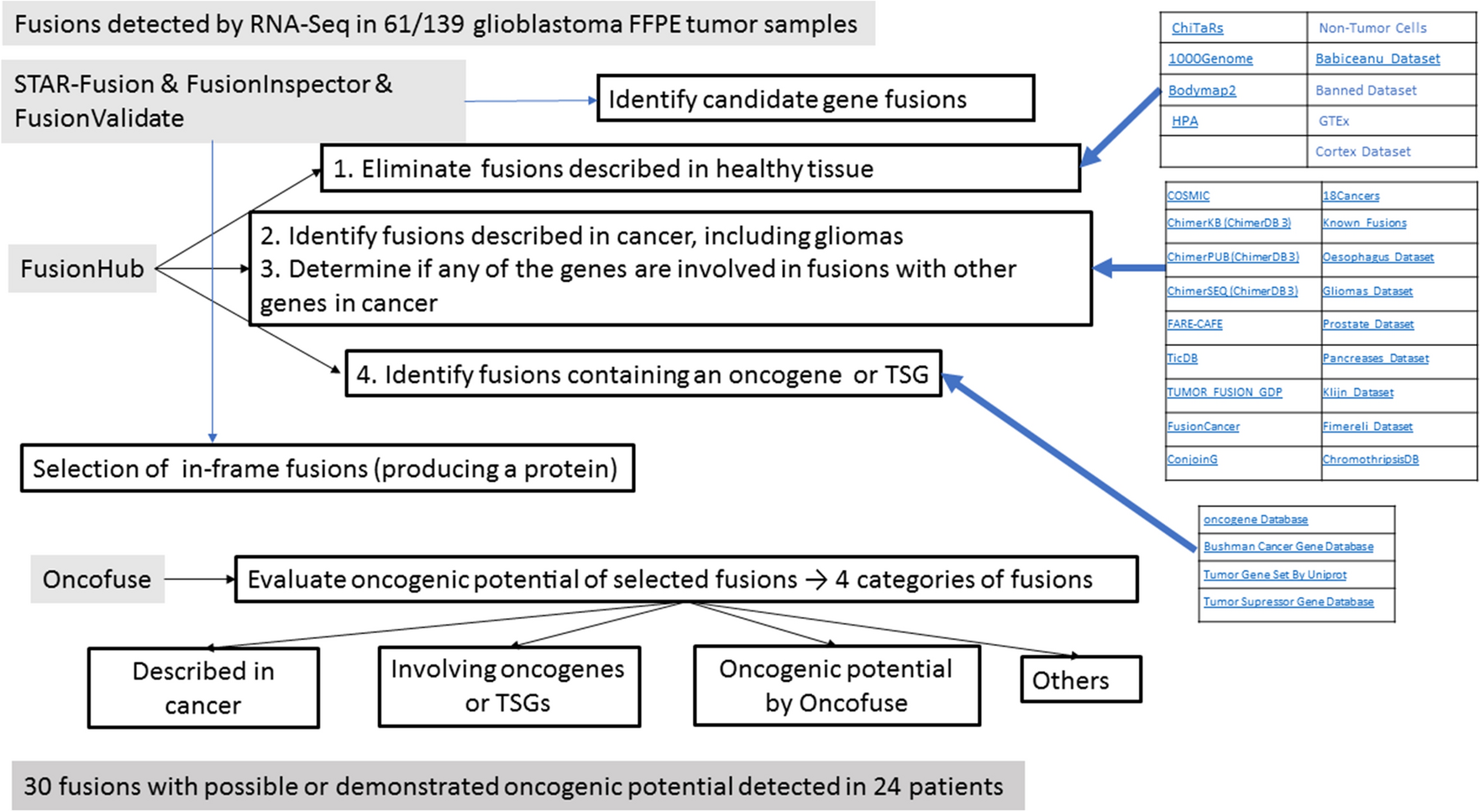
RNA-Sequencing (RNA-Seq) can identify gene fusions in tumors, but not all these fusions have functional consequences. Using multiple data bases, researchers at the Institut Catala d’Oncologia, Spain have performed an in silico analysis of fusions detected by RNA-Seq in tumor samples from 139 newly diagnosed glioblastoma patients to identify in-frame fusions with predictable oncogenic potential. Among 61 samples with fusions, there were 103 different fusions, involving 167 different genes, including 20 known oncogenes or tumor suppressor genes (TSGs), 16 associated with cancer but not oncogenes or TSGs, and 32 not associated with cancer but previously shown to be involved in fusions in gliomas. After selecting in-frame fusions able to produce a protein product and running Oncofuse, the researchers identified 30 fusions with predictable oncogenic potential and classified them into four non-overlapping categories: six previously described in cancer; six involving an oncogene or TSG; four predicted by Oncofuse to have oncogenic potential; and 14 other in-frame fusions. Only 24 patients harbored one or more of these 30 fusions, and only two fusions were present in more than one patient: FGFR3::TACC3 and EGFR::SEPTIN14. This in silico study provides a good starting point for the identification of gene fusions with functional consequences in the pathogenesis or treatment of glioblastoma.
Procedures and data bases used in the present study
to select the fusions with oncogenic potential
From the long list of fusions detected by RNA-Seq, we used STAR-Fusion to detect fusion genes and FusionInspector to validate predicted fusions. We then used FusionHub to eliminate fusions previously described in healthy tissue, identify fusions previously described in cancers, and explore whether either gene had been identified as an oncogene or tumor suppressor gene (TSG) or had been associated with cancer. We next used FusionValidate to select only in-frame fusions and finally ran Oncofuse to predict the oncogenic potential of each fusion.
Hernandez A, Muñoz-Mármol AM, Esteve-Codina A, Alameda F, Carrato C, Pineda E, Arpí-Lluciá O, Martinez-García M, Mallo M, Gut M, Del Barco S, Gallego O, Dabad M, Mesia C, Bellosillo B, Domenech M, Vidal N, Aldecoa I, de la Iglesia N, Balana C. (2022) In silico validation of RNA-Seq results can identify gene fusions with oncogenic potential in glioblastoma. Sci Rep 12(1):14439. [article]