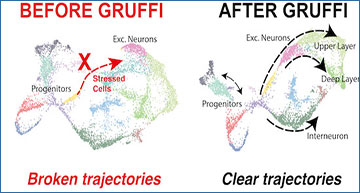
Organoids enable in vitro modeling of complex developmental processes and disease pathologies. Like most 3D cultures, organoids lack sufficient oxygen supply and therefore experience cellular stress. These negative effects are particularly prominent in complex models, such as brain organoids, and can affect lineage commitment. Researchers at the Institute of Molecular Biotechnology, Austria analyzed brain organoid and fetal single-cell RNA sequencing (scRNAseq) data from published and new datasets, totaling about 190,000 cells. They identified a unique stress signature in the data from all organoid samples, but not in fetal samples. The researchers demonstrated that cell stress is limited to a defined subpopulation of cells that is unique to organoids and does not affect neuronal specification or maturation. They have developed a computational algorithm, Gruffi, which uses granular functional filtering to identify and remove stressed cells from any organoid scRNAseq dataset in an unbiased manner. The researchers validated their method using six additional datasets from different organoid protocols and early brains, and show its usefulness to other organoid systems including retinal organoids. These data show that the adverse effects of cell stress can be corrected by bioinformatic analysis for improved delineation of developmental trajectories and resemblance to in vivo data.
Granular functional filtering identifies stressed cells unbiasedly
A. Gene-set scores per cell for the two strongest stress signatures (ER-stress and glycolysis) and the two cardinal processes in the developing brain (neurogenesis and gliogenesis). The complementary expression signatures suggest a mutually exclusive neural- or stressed- fate. B. Overview of Gruffi’s stress classification. After preprocessing steps including the computation of PCA and UMAP embeddings, a gene ontology pathway is selected, respective gene sets are retrieved, and per cell GO-scores are calculated. At the same time, an ideal clustering resolution is estimated, such that cells are assigned to granules of (in median) ~ 100–200 cells, and small clusters (< 30) are reclassified. Next, to overcome high variability and detection noise caused by single-cell resolution, average and cell number normalized granule scores are calculated, and respective score-thresholds are estimated based on the score’s dispersion. Finally, stressed granules are identified by a combination of scores, and isolated from the dataset for separate analysis or dataset cleaning and further downstream analysis is possible. C. Gruffi defined 995 granules by snn-clustering containing a median of 156 cells. D. Granule scores for glycolysis shown on UMAP. E. Three-dimensional stress score threshold estimation by Gruffi using default setting, requiring high glycolytic and ER-stress, low gliogenesis score to define stressed cells. F. Stressed cells classified by Gruffi based on ER-stress, glycolysis and gliogenesis are highlighted on the UMAP.
Availability – The Gruffi package is made available under github.com/jn-goe/gruffi. The code for analysis will be accessible on Github: github.com/vertesy/Limited.Stress.in.Brain.organoids.
Vértesy A, Eichmüller OL, Naas J et al. (2022) Gruffi: an algorithm for computational removal of stressed cells from brain organoid transcriptomic datasets. The EMBO Journal [Epub ahead of print]. [article]