The increasing application of RNA sequencing to study non-model species demands easy-to-use and efficient bioinformatics tools to help researchers quickly uncover biological and functional insights. Researchers at McGill University have developed ExpressAnalyst, a web-based platform for processing, analyzing, and interpreting RNA-sequencing data from any eukaryotic species. ExpressAnalyst contains a series of modules that cover from processing and annotation of FASTQ files to statistical and functional analysis of count tables or gene lists. All modules are integrated with EcoOmicsDB, an ortholog database that enables comprehensive analysis for species without a reference transcriptome. By coupling ultra-fast read mapping algorithms with high-resolution ortholog databases through a user-friendly web interface, ExpressAnalyst allows researchers to obtain global expression profiles and gene-level insights from raw RNA-sequencing reads within 24 h. Here, the researchers present ExpressAnalyst and demonstrate its utility with a case study of RNA-sequencing data from multiple non-model salamander species, including two that do not have a reference transcriptome.
Concept for and implementation of ExpressAnalyst
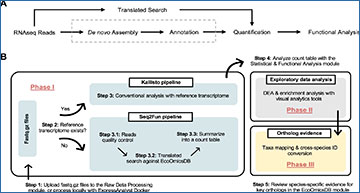
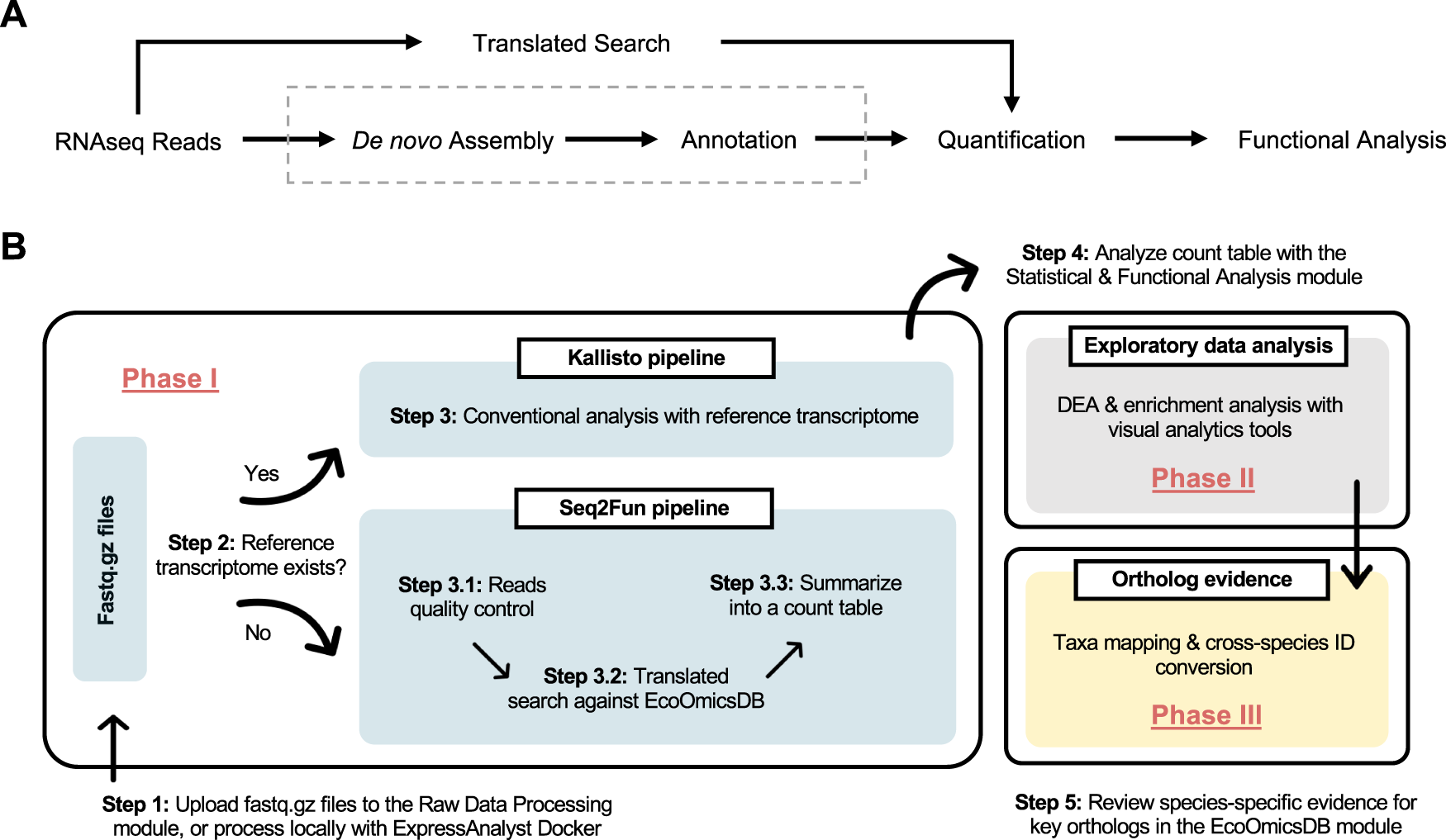
A Conceptual solution to bypass the computationally intensive steps of de novo assembly and annotation. B Practical implementation of the conceptual solution in ExpressAnalyst. DEA Differential expression analysis.
Availability – ExpressAnalyst is available at: https://www.expressanalyst.ca/
Liu P, Ewald J, Pang Z, Legrand E, Jeon YS, Sangiovanni J, Hacariz O, Zhou G, Head JA, Basu N, Xia J. (2023) ExpressAnalyst: A unified platform for RNA-sequencing analysis in non-model species. Nat Commun 14(1):2995. [article]