Single-cell RNA sequencing (scRNA-seq) is a powerful tool, unlocking the secrets of cellular heterogeneity, trajectory inference, and the identification of rare cell types. However, the high dimensionality, sparsity, and presence of ”false” zero values in the data can pose challenges to clustering. Furthermore, current unsupervised clustering algorithms have not effectively leveraged prior biological knowledge, making cell clustering even more challenging
Researchers at Shenzhen University have developed a novel semi-supervised clustering model dubbed scTPC. Unlike traditional clustering algorithms, scTPC harnesses the power of deep learning and integrates prior biological knowledge to navigate the complexities of scRNA-seq data.
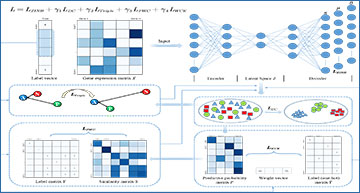
At the heart of scTPC lies a denoising autoencoder, pre-trained on a zero-inflated negative binomial (ZINB) distribution—a crucial step to mitigate the high dimensionality, sparsity, and “false” zero values inherent in scRNA-seq datasets. Building upon this foundation, scTPC embarks on deep clustering in the learned latent feature space, guided by triplet and pairwise constraints derived from partially labeled cells.
What sets scTPC apart is its ability to address the inherent imbalances in cell-type datasets. By introducing a weighted cross-entropy loss, the model optimizes its performance, ensuring accurate clustering even in the face of skewed data distributions.
The efficacy of scTPC is demonstrated through a series of experiments on both real and simulated scRNA-seq datasets. Across ten real datasets and five simulated ones, scTPC consistently delivers precise clustering results, unveiling the intricate tapestry of cellular diversity with its meticulously designed framework.
With its ability to harness the power of deep learning and integrate prior knowledge, scTPC represents a significant advancement in scRNA-seq data analysis—an invaluable asset in unraveling the mysteries of the cellular world.
Availability – scTPC is a Python-based algorithm, and the code is available from https://github.com/LF-Yang/Code or https://zenodo.org/records/10951780.
Qiu Y, Yang L, Jiang H, Zou Q. (2024) scTPC: a novel semi-supervised deep clustering model for scRNA-seq data. Bioinformatics [Epub ahead of print]. [abstract]