The advent of next-generation sequencing (NGS) technologies has revolutionized the way researchers study genetic information. However, despite its remarkable capabilities, NGS platforms are not without limitations. One significant challenge lies in the sequencing error rate, which stands at 1 per 1000 base pairs (10^-3). This error rate poses a substantial obstacle, particularly in fields where precise detection of rare mutations is crucial, such as somatic mosaicism or microbial heterogeneity.
A recent study delves into this issue, examining the development of high-fidelity sequencing methods over the past decade. The researchers identify three major factors contributing to sequencing errors and propose twelve strategies to mitigate these errors effectively. These strategies encompass a range of approaches, from refining sequencing chemistry to optimizing data analysis algorithms.
To provide a comprehensive overview, the study categorizes eleven existing high-fidelity sequencing methods based on their underlying error-reduction strategies. This classification framework highlights common themes and allows for a systematic comparison of these approaches. Additionally, the researchers identify three key trends that have emerged in the evolution of sequencing methodologies, shedding light on the direction of future advancements.
Causes underlying sequencing errors and strategies reducing errors
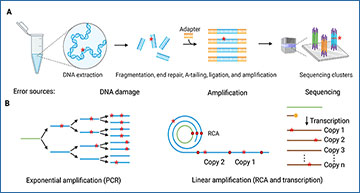
A. An overall schema of the typical Illumina sequencing process. B. Two types of amplification modes. C. Error avoidance during end repair or A-tailing. D. The sequencing error distribution of Illumina paired-end sequencing (2 × 150 bp) in libraries with short and long insert sizes. E. Mutation signature with or without removal of DNA damage. F. Strategies for reducing sequencing errors
Expanding their analysis beyond traditional short-read sequencing methods, the researchers also explore eight long-read sequencing techniques. Long-read sequencing offers distinct advantages, including the ability to span complex genomic regions with high accuracy. The study underscores the importance of error reduction strategies in long-read sequencing and identifies areas for further improvement.
Looking ahead, the study proposes two promising avenues for future research. These include the development of novel sequencing chemistries and the integration of machine learning algorithms to enhance accuracy while reducing costs. By leveraging advancements in technology and methodology, researchers aim to push the boundaries of NGS and long-read sequencing, unlocking new insights into the complexities of the genome.
This study underscores the importance of addressing sequencing errors in NGS and long-read sequencing technologies. By implementing innovative strategies and embracing emerging trends, researchers can overcome existing challenges and propel genomic research forward into new frontiers of discovery.
Jia H, Tan S, Zhang YE. (2024) Chasing Sequencing Perfection: Marching Toward Higher Accuracy and Lower Costs. GPB [Epub ahead of print]. [abstract]