In cancer research, acquiring diverse and large datasets is often challenging and costly. However, a groundbreaking study by researchers at Stanford University School of Medicine has introduced a novel approach to overcome this hurdle: using synthetically generated data to train machine-learning models. Cascaded diffusion models are revolutionizing cancer research by synthesizing realistic whole-slide image tiles from RNA-sequencing data, paving the way for accelerated model development and data imputation.
The study employs cascaded diffusion models to create synthetic whole-slide image tiles from latent representations of RNA-sequencing data obtained from human tumors. These models are trained to generate images that accurately reflect the cellular composition and gene expression alterations observed in bulk RNA-sequencing data across various cancer types, including lung adenocarcinoma, kidney renal papillary cell carcinoma, cervical squamous cell carcinoma, colon adenocarcinoma, and glioblastoma.
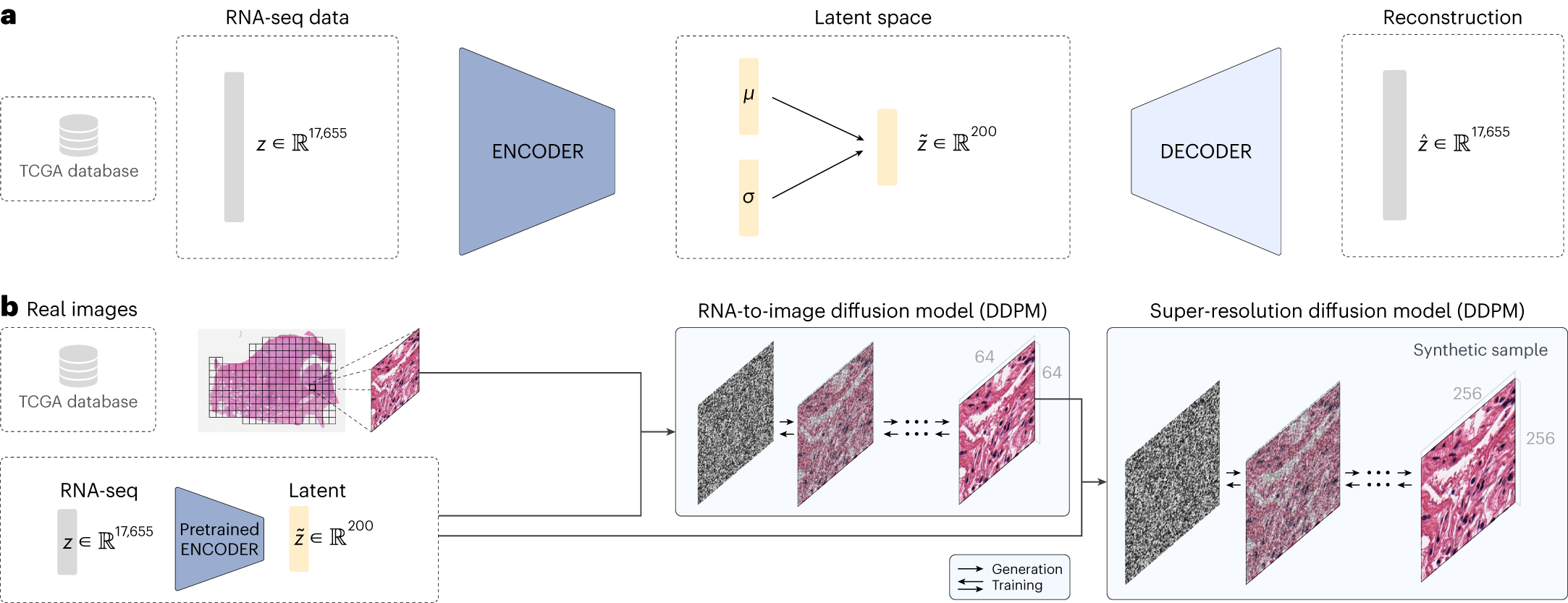
RNA-CDM model architecture used for generating RNA-seq embeddings
and synthetic WSI tiles using diffusion models
a, Beta-VAE architecture for the generation of gene-expression embeddings. The model uses as input the expression of 17,655 genes. Both the encoder and the decoder are formed by two linear layers of 6,000 and 4,000, respectively. The latent μ and σ vectors have a feature size of 200. b, RNA-CDM architecture for generating synthetic multicancer tiles. It is formed by two Denoising Diffusion Probabilistic Models (DDPM), one acting as an RNA-to-image model and the second as a super-resolution model. The pretrained encoder is used to obtain the latent representation of the RNA-seq data. A corresponding tile from the patient is obtained. For the first DDPM, the tile is resized from 256 × 256 to 64 × 64 pixels. During the training phase, noise is gradually applied to the tile according to the given noise scheduler at each timestep t. Then, the first DDPM learns to reduce the noise by using as input the noisy image, the timestep t and the gene-expression embedding. The noise predicted is removed from the noisy image at each timestep, having a denoised tile of 64 × 64 at the end of the process. Then, a second DDPM takes the denoised image, the noisy 256 × 256 image at timestep t, the timestep t and the gene-expression embedding, and predicts the added noise again. Then, the noise is removed from the 256 × 256 tile iteratively until a denoised image is obtained and compared with the original tile. For generating a new image, the process is the same, but we start from total random noise until we have a synthetic tile whose generation has been guided by the gene-expression embedding.
One of the key findings of the study is that alterations in gene expression directly influence the composition of cell types in the synthetic image tiles. Despite being generated synthetically, these images faithfully preserve the distribution of cell types and maintain the cell fraction observed in bulk RNA-sequencing data. This breakthrough demonstrates the potential of synthetic data in overcoming data scarcity challenges in cancer research.
Furthermore, machine-learning models pretrained with the generated synthetic data exhibit superior performance compared to models trained from scratch. By leveraging synthetic data for training, researchers can develop more robust and accurate machine-learning models even in settings where data availability is limited. This not only accelerates the pace of model development but also enhances the reliability of predictive analytics in cancer research.
The use of synthetic data holds immense promise in accelerating cancer research and facilitating advancements in precision medicine. By providing researchers with access to diverse and realistic datasets, synthetic data can expedite the development of machine-learning models for cancer diagnosis, prognosis, and treatment prediction. Additionally, synthetic data enables the imputation of missing data modalities, further enhancing the comprehensiveness of analytical models.
Moreover, the adoption of synthetic data approaches democratizes access to advanced machine-learning techniques, leveling the playing field for researchers worldwide. By reducing reliance on costly and resource-intensive data acquisition processes, synthetic data empowers researchers to explore new avenues of inquiry and drive innovation in cancer biology and therapeutics.
The integration of cascaded diffusion models for synthesizing realistic data represents a paradigm shift in cancer research. By harnessing the power of synthetic data, researchers can overcome the limitations of data scarcity and accelerate the development of machine-learning models for cancer analysis. This transformative approach holds the potential to revolutionize cancer research, ultimately leading to improved patient outcomes and advancements in personalized medicine.
Carrillo-Perez F, Pizurica M, Zheng Y, Nandi TN, Madduri R, Shen J, Gevaert O. (2024) Generation of synthetic whole-slide image tiles of tumours from RNA-sequencing data via cascaded diffusion models. Nat Biomed Eng [Epub ahead of print]. [abstract]