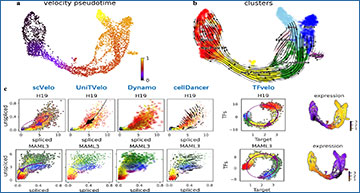
Understanding the dynamics of gene expression within single cells is crucial for deciphering cell fate and predicting cellular states. A recent study introduces a novel concept called RNA velocity, which offers insights into the future trajectory of individual cells based on their RNA abundance data. However, existing RNA velocity models face challenges in accurately capturing gene dynamics. To address this issue, researchers at Shanghai Jiao Tong University have developed TFvelo, a groundbreaking approach that expands the RNA velocity concept by incorporating gene regulatory information.
Traditionally, RNA velocity models rely on the phase delay between unspliced and spliced mRNA to infer cell dynamics. However, this approach may not always provide sufficient signal for dynamic modeling, leading to inaccuracies in predicting cell trajectories. Motivated by the idea that RNA velocity could be influenced by transcriptional regulation, TFvelo takes a different approach by integrating gene regulatory information into its analysis.
In essence, TFvelo aims to accurately model RNA velocity across various single-cell datasets without solely relying on splicing information. By introducing gene regulatory data, TFvelo can effectively capture gene dynamics on phase portraits, providing a more accurate representation of cell trajectories and pseudo-time inference.
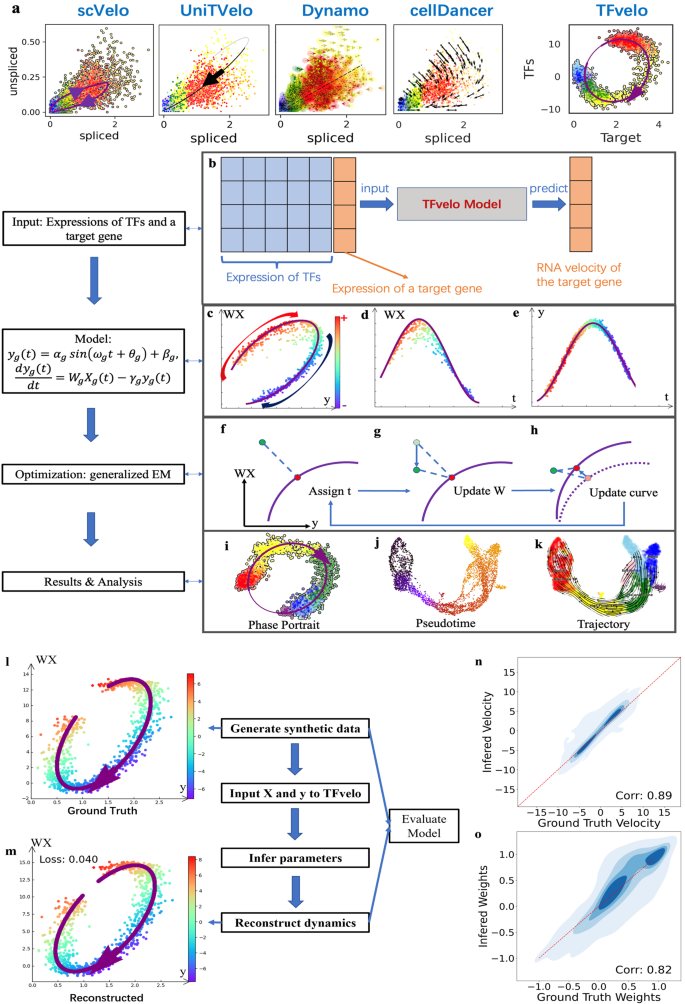
The overview of TFvelo
a The comparison among constructed dynamics on LITAF gene in Pancreas dataset by TFvelo and previous approaches. b The workflow of TFvelo. c–e The phase portrait plot on WX-y space, the plot of WX with respect to pseudotime, and the plot of y with respect to pseudotime. f–h The steps within each iteration of the generalized EM algorithm. f Assignment of latent time to each cell (in green) to find the corresponding target point (shown in red), which is the nearest point to the cell on the purple line. g Optimization of the weights to move all cells along the axis, minimizing the mean loss over all cells. h Optimization of the parameters in the dynamical function, to move the target point on the curve closer to each cell. i–k Visualization and analysis of TFvelo’s results, including phase portrait fitting pseudo-time inference, and cell trajectory prediction. l–o Simulation on synthetic dataset. l An example of the generated synthetic dynamics. m Reconstructed dynamics by TFvelo of the sample shown in (l) with MSE loss. n The joint plot of ground truth velocity and inferred velocity with their spearman correlation. The red reference line refers to that the ground truth is equal to the inferred value. o The joint plot of ground truth weights and inferred weights with their spearman correlation.
To validate the effectiveness of TFvelo, researchers conducted experiments on synthetic data and multiple single-cell RNA sequencing datasets. The results demonstrated that TFvelo outperformed existing RNA velocity models, accurately fitting gene dynamics and inferring cell pseudo-time and trajectory from RNA abundance data.
Overall, TFvelo represents a robust and accurate avenue for modeling RNA velocity in single-cell data. By incorporating gene regulatory information, TFvelo offers a more comprehensive understanding of gene dynamics within individual cells, paving the way for advancements in cell fate prediction and cellular state analysis. With further validation and application, TFvelo holds great potential for unraveling the complexities of cellular dynamics and enhancing our understanding of biological systems at the single-cell level.
Availability – TFvelo is implemented in Python, based on the scVelo package. The source code can be downloaded from the GitHub repository, https://github.com/xiaoyeye/TFvelo.
Li J, Pan X, Yuan Y, Shen HB. (2024) TFvelo: gene regulation inspired RNA velocity estimation. Nat Commun 15(1):1387. [article]