Long-read RNA sequencing has emerged as a powerful tool for transcript discovery, even in well-annotated organisms. However, assessing the accuracy of different methods in identifying annotated and novel transcripts remains a challenge. Researchers at the Spanish National Research Council have developed SQANTI-SIM, a versatile tool that wraps around popular long-read simulators to allow precise management of transcript novelty based on the structural categories defined by SQANTI3. By selectively excluding specific transcripts from the reference dataset, SQANTI-SIM effectively emulates scenarios involving unannotated transcripts. Furthermore, the tool provides customizable features and supports the simulation of additional types of data, representing the first multi-omics simulation tool for the lrRNA-seq field.
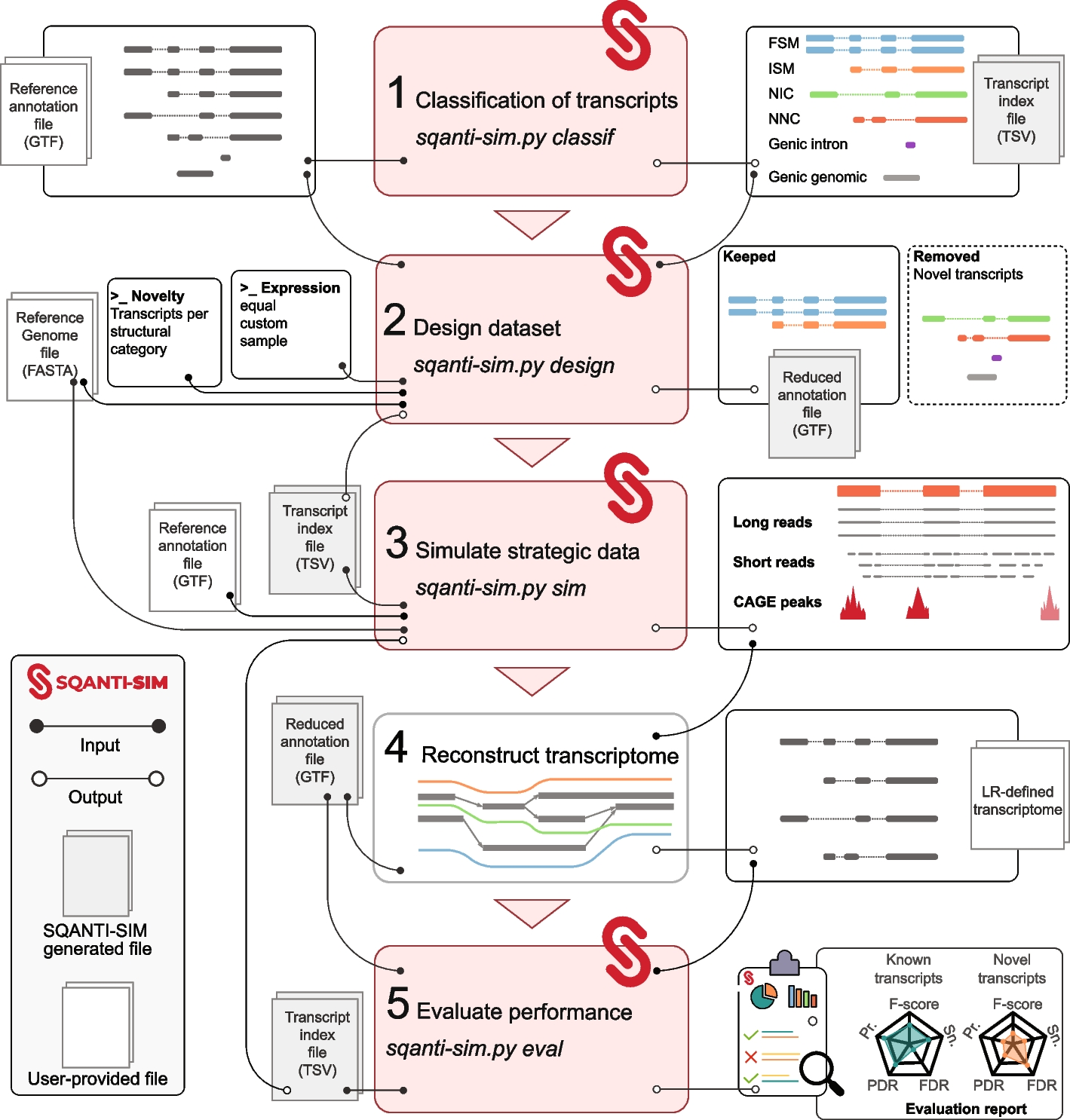
Flowchart of the SQANTI-SIM pipeline
The first three steps simulate reads and accompanying datasets according to the user’s specifications. Simulated data is then used by the transcriptome reconstruction algorithm to predict transcripts. The last SQANTI-SIM module assesses performance by comparison to the simulated ground truth and provides a comprehensive evaluation report
Availability – SQANTI-SIM is available at https://github.com/ConesaLab/SQANTI-SIM
Mestre-Tomás J, Liu T, Pardo-Palacios F, Conesa A. (2023) SQANTI-SIM: a simulator of controlled transcript novelty for lrRNA-seq benchmark. Genome Biol 24(1):286. [article]