Transcription, the process of synthesizing RNA from DNA, lies at the heart of molecular biology and holds immense potential for applications in mRNA vaccines and RNA therapeutics. However, current methods for characterizing RNA suffer from biases introduced during amplification and enzymatic processes, leading to the loss of crucial native information. In a groundbreaking study, researchers at the University of Cambridge have introduced a novel strategy that harnesses the power of RNA nanotechnology and nanopores to quantitatively study transcription and RNA polymerase behavior.
The researchers employed T7 RNA polymerase to transcribe linear DNA sequences lacking termination signals. Surprisingly, they stumbled upon alternative transcription termination sites within the origin of replication sequence, uncovering new insights into the transcription process. Building on this discovery, the researchers turned their attention to circular DNA templates devoid of transcription terminators. Through a technique known as rolling circle transcription, they were able to delve deeper into the processivity and behavior of RNA polymerase at the single-molecule level.
RNA identifier (ID) assembly enables single-molecule
structural analysis of RNA transcripts using nanopore sensing
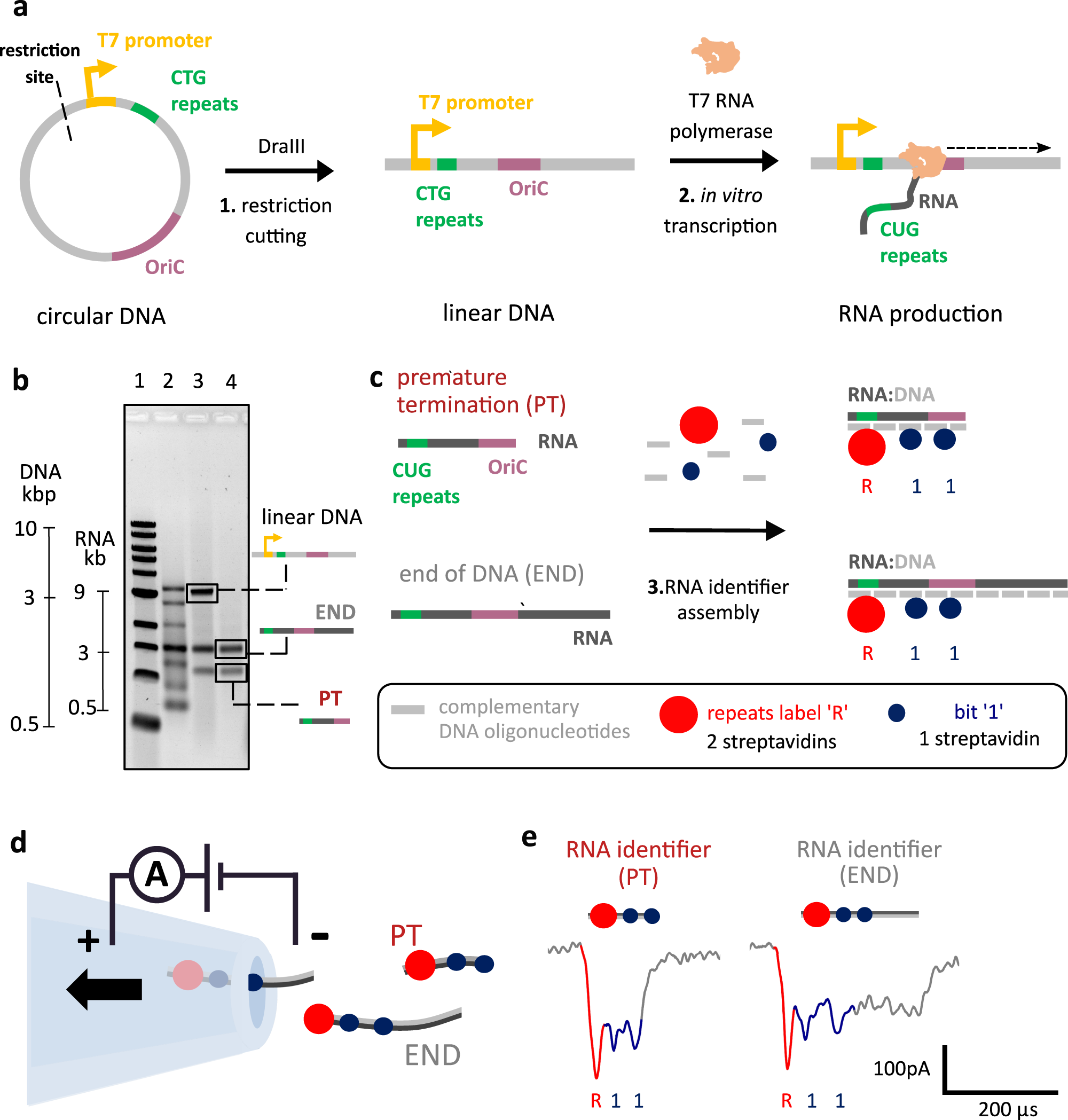
a In vitro transcription of linear DNA containing 12 CTG tandem repeats. A circular DNA construct contains a single T7RNAP promoter, the OriC, 12 CTG tandem repeats, and a DraIII restriction site. The circular DNA construct was linearized using DraIII, by cutting upstream of the T7RNAP promoter. The linear DNA was in vitro transcribed using T7RNAP. b Agarose gel electrophoresis of transcribed linear DNA indicates the presence of two RNA species (lane 4), one in which the RNA polymerase transcribes the entire linear DNA (END) and RNA of unknown nature. With nanopore sensing, it is later revealed that the lower band corresponds to RNA, where transcription is prematurely terminated within the OriC sequence (PT). Gel lanes: 1 – 1 kbp ladder; 2 – single-stranded RNA ladder; 3 – transcribed RNA from linear DNA; 4 – transcribed RNA from linear DNA treated with DNase I. Transcriptions were performed in triplicate. c RNA transcripts were hybridized with short complementary DNA oligonucleotides (~40 nt), producing RNA IDs. CUG repeats in RNA were labeled with two streptavidins (repeats label ”R”), indicating the beginning of transcription given their vicinity to the promoter. Further positioning along the transcript is achieved using bits “1” that have one streptavidin enabling their distinction from label “R”. d RNA IDs made from RNA transcripts that have different termination points (PT or END) translocating through the nanopore. Nanopore readout is based on electrophoretically driven transport of negatively charged RNA IDs through the nanopore towards a positively charged electrode. e Nanopore RNA ID readouts for PT and END RNAs are presented left and right, respectively. RNA ID for both PT and END shows downward spikes associated to the labeled repeats “R” (red) and “1” bits (blue). PT translocation finishes right after the second “1” bit hence making a clear distinction towards END translocations, which takes longer to translocate through the pore. Source data are provided as a Source Data file.
The study yielded remarkable findings, shedding light on the intricacies of RNA transcription. By leveraging RNA nanotechnology and nanopores, researchers gained unprecedented insights into RNA polymerase behavior and transcription dynamics. The novel approach enabled the direct and quantitative analysis of RNA transcripts, overcoming the limitations of existing methods. Notably, the methodology opens up exciting possibilities for accurate RNA structural mapping by facilitating the study of full-length RNA molecules at the single-molecule level.
This groundbreaking study represents a significant leap forward in our understanding of RNA transcription. By harnessing the combined power of RNA nanotechnology and nanopores, researchers have unlocked new avenues for unraveling the mysteries of RNA synthesis. The methodology pioneered in this study holds immense promise for advancing our knowledge of RNA biology and has far-reaching implications for fields such as drug development and molecular diagnostics. As scientists continue to explore the potential of this innovative approach, we can anticipate further breakthroughs that will revolutionize our understanding of transcription and RNA-based therapeutics.
Patiño-Guillén G, Pešović J, Panić M, Savić-Pavićević D, Bošković F, Keyser UF. (2024) Single-molecule RNA sizing enables quantitative analysis of alternative transcription termination. Nat Commun 15(1):1699. [article]