Estimating RNA modifications from Nanopore direct RNA sequencing data is an important task for the RNA research community. Current computational methods could not provide satisfactory results due to the inaccurate segmentation of the raw signal. Researchers at Aalto University have developed a new method, SegPore, that utilizes a molecular jiggling translocation hypothesis to segment the raw signal. SegPore is a pure white-box model with a superior interpretability, which significantly reduces structured noise in the raw signal. Based on the improved signal segmentation, SegPore+m6Anet has achieved state-of-the-art performance in m6A identification. Additionally, the researchers demonstrate SegPore’s interpretable results and decent performances on inosine modification estimation and RNA secondary structure estimation. An interesting discovery in RNA structure estimation is that the end points of the reads take place at the start of stem structures along the reverse transcription direction. These results indicate SegPore’s capability to concurrently estimate multiple modifications at the individual molecule level from the same Nanopore direct RNA sequencing data, as well as shed light on RNA structure estimation from a novel angle.
SegPore workflow
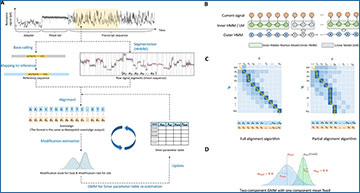
(A) The general workflow. First, basecalling and mapping are performed using Guppy and Minimap2 such that a raw current signal fragment is paired with a reference sequence fragment. Meanwhile, the raw current signal of a read is split into segments by HHMM and an estimated mean (𝜇𝑖 ) is derived for each segment. Then, the current signal segments ( 𝜎𝑖𝑖 ) are aligned with the 5mer list of the corresponding reference sequence fragment using the full/partial alignment algorithm, given a 5mer parameter table. Here we use A𝑗 to denote A at 𝑗𝑗th position on the reference. Next, all aligned to the same 5mer at different genomic locations are pooled together and a two-component GMM is fitted to re-estimate the 5mer parameters. One GMM component models the unmodified state and the other models the modified state, while the hidden variable of the GMM specifies the modification state of the 5mer on each read. The parameter estimation process is iterated several times on the training data to gain a stable estimation of the 5mer parameter table. The final 5mer parameter table is used for estimating the modification states on the test data. (B) Hierarchical hidden Markov model. The outer HMM partitions current signal into alternating base blocks and transition blocks. An inner HMM approximates the emission probability of a base block by considering neighboring 5mers. A linear model approximates the emission probability of a transition block. (C) Full/partial alignment algorithms. Each row is an estimated mean of a base block given by HHMM. Each column is a 5mer of the reference sequence. One 5mer can be aligned to multiple means. (D) Gaussian mixture model (GMM) for estimating modification states. The green component codes the unmodified state of a given 5mer. The blue component codes the modified state of the given 5mer. Each component has three parameters: mean (𝜇𝜇), std (𝜎𝜎) and weight (𝜔𝜔).
Availability – The source code is hosted on GitHub (https://github.com/guangzhaocs/SegPore).
Cheng G, Vehtari A, Cheng L. (2024) Raw signal segmentation for estimating RNA modifications and structures from Nanopore direct RNA sequencing data. bioRXiv [Epub ahead of print]. [article]