Understanding the cellular basis of disease phenotypes is crucial for developing targeted therapies. Traditional bulk assays like RNA sequencing have limitations in identifying specific groups of cells driving disease phenotypes, especially when marker genes are unknown or detectable only at later stages. However, recent advancements in single-cell RNA sequencing (scRNA-seq) offer a revolutionary approach by providing gene expression profiles at the resolution of individual cells.
The Promise of ScRAT
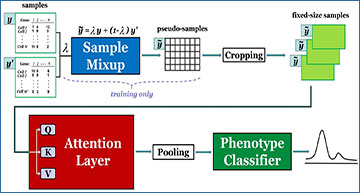
Researchers at Simon Fraser University have developed ScRAT, which stands for Single-Cell RNA-seq-based Phenotype Prediction Tool, a new method that addresses the challenge of identifying disease-driving cells with limited annotated samples. Unlike existing methods, ScRAT does not rely on accurate cell type annotations and can effectively predict phenotypes, such as coronavirus disease (COVID), even with a small number of training samples. The key innovation of ScRAT lies in its utilization of a mixup module to augment the training dataset and a multi-head attention mechanism to identify informative cells associated with each phenotype.
An overview of ScRAT
ScRAT consists of three main modules: Aample Mixup, Attention Layer, and Phenotype Classifier. It takes a scRNA-seq sample (a set of cells) as input, and outputs the predicted phenotype for the input sample.
Performance and Validation
To assess the efficacy of ScRAT, the researchers tested it on three public COVID datasets and compared its performance with other phenotype prediction methods. Remarkably, ScRAT outperformed its competitors, demonstrating superior accuracy even with a reduced number of training samples. Moreover, ScRAT’s ability to identify critical cell types based on high-attention cells corroborated findings from original studies and recent literature, suggesting its potential for uncovering novel molecular mechanisms and therapeutic targets.
Implications for Precision Medicine
The development of ScRAT represents a significant advancement in our ability to unravel disease phenotypes using scRNA-seq data. By overcoming the limitations of traditional methods and leveraging deep learning techniques, ScRAT offers a powerful tool for identifying disease-driving cells and understanding the underlying molecular mechanisms. This has profound implications for precision medicine, as it enables the development of targeted therapies tailored to the specific cellular components driving disease pathogenesis.
As we continue to unravel the complexities of disease at the single-cell level, innovative tools like ScRAT hold immense promise for advancing our understanding of disease phenotypes and facilitating the development of personalized treatment strategies. By harnessing the power of scRNA-seq data and deep learning algorithms, ScRAT represents a pivotal step towards realizing the full potential of precision medicine in the fight against various diseases, including COVID-19 and beyond.
Availability: The code of the proposed method ScRAT is published at https://github.com/yuzhenmao/ScRAT.
Mao Y, Lin YY, Wong NKY, Volik S, Sar F, Collins C, Ester M. (2024) Phenotype prediction from single-cell RNA-seq data using attention-based neural networks. Bioinformatics 40(2):btae067. [article]