RNA G-quadruplexes (rG4s) are non-canonical structural motifs that have diverse functional and regulatory roles, for instance in transcription termination, alternative splicing, mRNA localization and stabilization, and translational process. Researchers at the City University of Hong Kong recently developed the RNA G-quadruplex structure sequencing (rG4-seq) technique and described rG4s in both eukaryotic and prokaryotic transcriptomes. However, rG4-seq suffers from a complicated gel purification step and limited PCR product yield, thus requiring a high amount of RNA input, which limits its applicability in more physiologically or clinically relevant studies often characterized by the limited availability of biological material and low RNA abundance. Here, they redesign and enhance the workflow of rG4-seq to address this issue.
The researchers developed rG4-seq 2.0 by introducing a new ssDNA adapter containing deoxyuridine during library preparation to enhance library quality with no gel purification step, less PCR amplification cycles and higher yield of PCR products. They demonstrate that rG4-seq 2.0 produces high-quality cDNA libraries that support reliable and reproducible rG4 identification at varying RNA inputs, including RNA mounts as low as 10 ng. rG4-seq 2.0 also improved the rG4-seq calling outcome and nucleotide bias in rG4 detection persistent in rG4-seq 1.0. The researchers further provide in vitro mapping of rG4 in the HEK293T cell line, and recommendations for assessing RNA input and sequencing depth for individual rG4 studies based on transcript abundance.
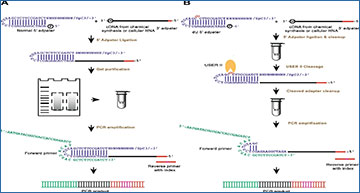
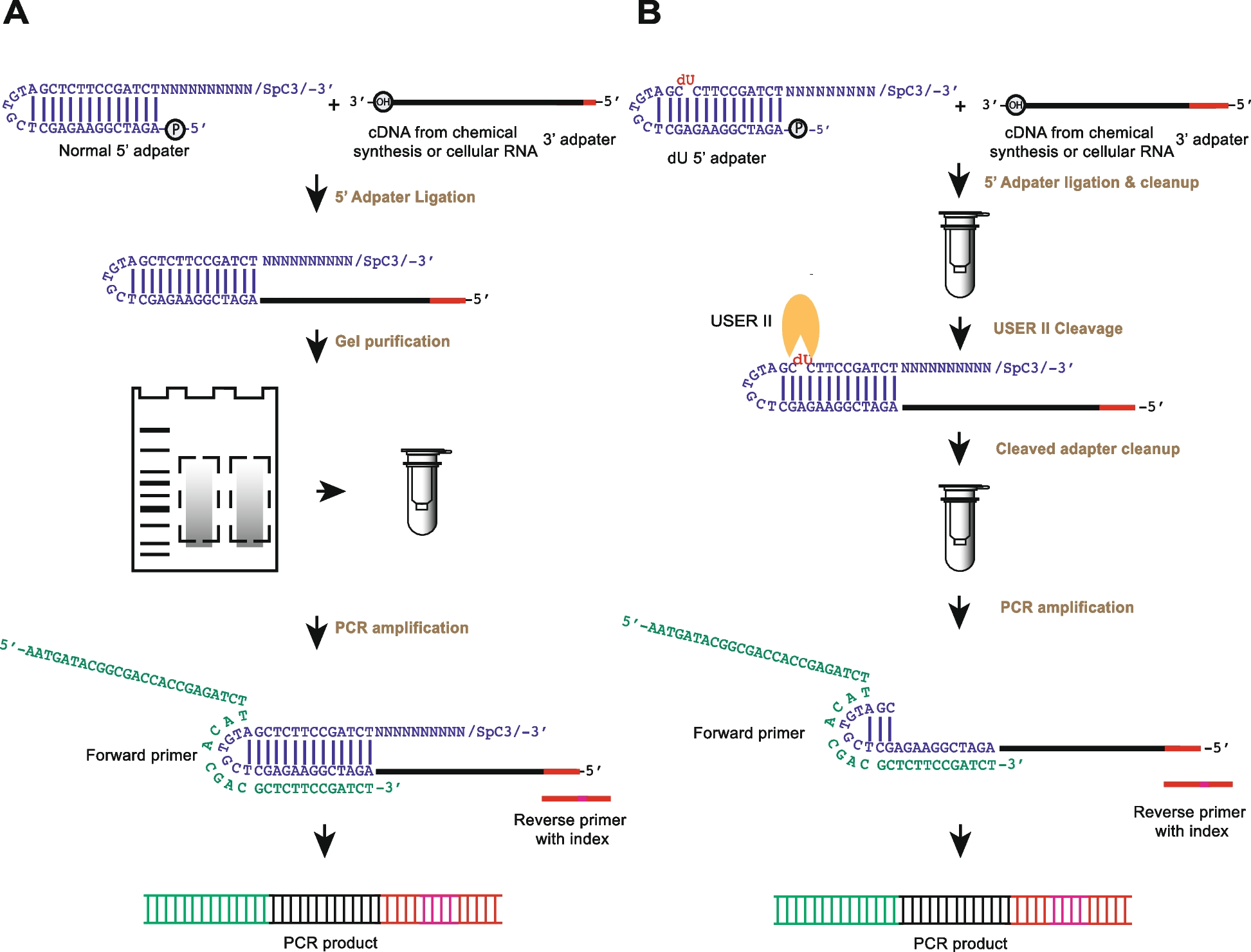
Comparison of the experimental strategy of rG4-seq and rG4-seq 2.0
from 5‘ adapter ligation to PCR amplification
The experimental pipelines that use A normal 5′ adapter and B 5′ dU adapter are shown (blue). In A, the normal 5′ adapter ligates to the cDNA (black) with 3′ adapter sequence (red) followed by gel purification before performing PCR amplification. The 6nt index region in the reverse PCR primer is in pink. In B, dU adapter is used for the ligation, and after the column purification, the enzymatic cleavage step is performed with cleavage enzyme (USER II, orange pacman) followed by column clean-up before the PCR amplification. USER II enzyme cleaves the 5′ adapter at the dU position as indicated in red in the 5′ dU adapter. The dU adapter contains the same secondary structure as the normal 5′ adapter and carries a 5′ phosphate at its 5′ end to ligate to incoming cDNA with a 3′hydroxyl, and a C3 spacer group at its 3′ end to block and avoid self-oligomerization. The 10 random nucleotides (N10) serve as a template for the hybridization of any incoming cDNA
Zhao J, Chow EY, Yeung PY, Zhang QC, Chan TF, Kwok CK. (2022) Enhanced transcriptome-wide RNA G-quadruplex sequencing for low RNA input samples with rG4-seq 2.0. BMC Biol 20(1):257. [article]