The ability to visualize RNA in its native subcellular environment by using single-molecule fluorescence in situ hybridization (smFISH) has reshaped our understanding of gene expression and cellular functions. A major hindrance of smFISH is the difficulty to perform systematic experiments in medium- or high-throughput formats, principally because of the high cost of generating the individual fluorescent probe sets.
Researchers at the University of Montpellier have developed high-throughput smFISH (HT-smFISH), a simple and cost-efficient method for imaging hundreds to thousands of single endogenous RNA molecules in 96-well plates. HT-smFISH uses RNA probes transcribed in vitro from a large pool of unlabeled oligonucleotides. This allows the generation of individual probes for many RNA species, replacing commercial DNA probe sets. HT-smFISH thus reduces costs per targeted RNA compared with many smFISH methods and is easily scalable and flexible in design. The researchers provide a protocol that combines oligo pool design, probe set generation, optimized hybridization conditions and guidelines for image acquisition and analysis. The pipeline requires knowledge of standard molecular biology tools, cell culture and fluorescence microscopy. It is achievable in ~20 d. In brief, HT-smFISH is tailored for medium- to high-throughput screens that image RNAs at single-molecule sensitivity.
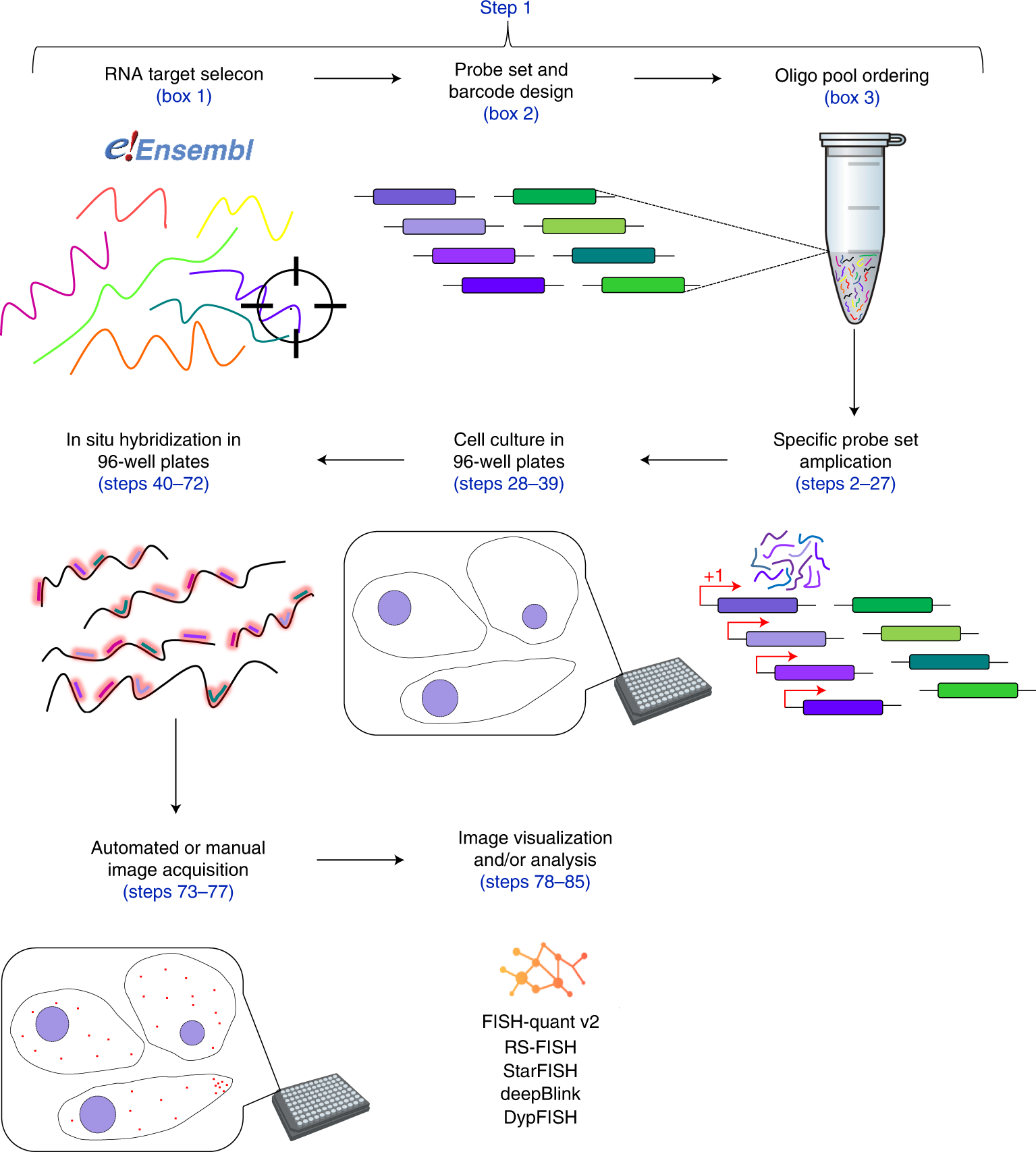
Overview of the HT-smFISH pipeline
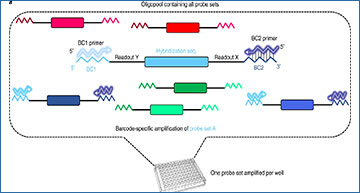
The pipeline starts with the desired RNA target selection and sequence extraction. Sequences then serve as input for a dedicated code (Oligostan-HT) used to generate thermodynamically optimized probe sets, as well as barcodes. Probe sets are then ordered as single-stranded DNA oligonucleotides in a custom oligo pool. These oligonucleotides are used to generate transcript-specific RNA probe sets via PCR amplification and IVT (Steps 2–27). For high-throughput RNA screening experiments, cells are grown in 96-well plates (Steps 28–39). In situ hybridization is also carried out (Steps 40–72) and imaged (Steps 73–77) directly on 96-well plates. Images can be directly visualized and/or analyzed with various tools such as FISH-quant v2, RS-FISH, StarFISH, deepBlink and DypFISH (Steps 78–85).
Safieddine A, Coleno E, Lionneton F, Traboulsi AM, Salloum S, Lecellier CH, Gostan T, Georget V, Hassen-Khodja C, Imbert A, Mueller F, Walter T, Peter M, Bertrand E. (2022) HT-smFISH: a cost-effective and flexible workflow for high-throughput single-molecule RNA imaging. Nat Protoc [Epub ahead of print]. [abstract]