Large compendia of gene expression data have proven valuable for the discovery of novel biological relationships. Historically, most available RNA assays were run on microarray, while RNA-seq is now the platform of choice for many new experiments. The data structure and distributions between the platforms differ, making it challenging to combine them directly.
Researchers at the University of Pennsylvania School of Medicine performed supervised and unsupervised machine learning evaluations to assess which existing normalization methods are best suited for combining microarray and RNA-seq data. They found that quantile and Training Distribution Matching normalization allow for supervised and unsupervised model training on microarray and RNA-seq data simultaneously. Nonparanormal normalization and z-scores are also appropriate for some applications, including pathway analysis with Pathway-Level Information Extractor (PLIER). The researchers demonstrate that it is possible to perform effective cross-platform normalization using existing methods to combine microarray and RNA-seq data for machine learning applications.
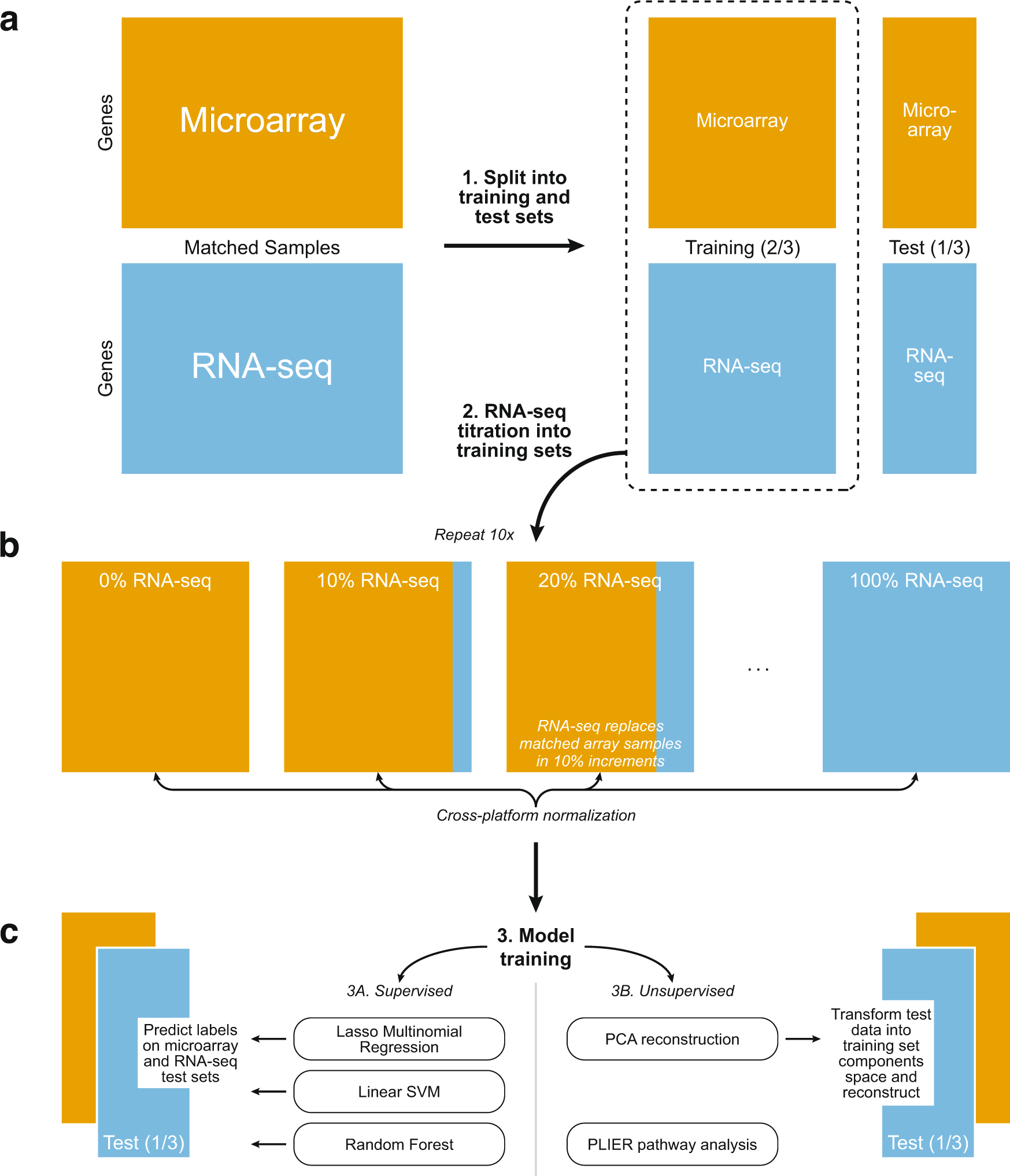
Overview of supervised and unsupervised machine learning experiments
a TCGA matched samples (520 from BRCA, 150 from GBM) run on both microarray and RNA-seq were split into a training set (2/3) and test set (1/3). b RNA-seq samples were titrated into each training set, 10% at a time (0–100%), resulting in eleven training sets for each normalization method. Each RNA-seq sample replaces its matched microarray sample. Cross-platform normalization methods were applied to each training set independently. c We used three supervised algorithms to train classifiers (molecular subtype and mutation status of TP53 and PIK3CA in both BRCA and GBM) on each training set and tested on the microarray and RNA-seq test sets. The test sets were projected onto and back out of the training set space using unsupervised Principal Components Analysis to obtain reconstructed test sets. The subtype classifiers trained in step 3A were used to predict on the reconstructed test sets. Pathways regulating gene expression were identified using the unsupervised method PLIER.
Foltz SM, Greene CS, Taroni JN. (2023) Cross-platform normalization enables machine learning model training on microarray and RNA-seq data simultaneously. Commun Biol 6(1):222. [article]