In the realm of molecular biology, understanding the intricate processes of RNA synthesis and turnover is paramount. A fascinating tool in this endeavor is the use of nucleoside analogues like 4-thiouridine (4sU), which allow researchers to label newly synthesized RNA molecules within cells.
In a recent study, scientists at the University of Regensburg delved into the effects of extensive 4sU labeling on RNA expression estimates using a technique called nucleotide conversion RNA-seq. This method involves chemically converting 4sU-labeled RNA into T-to-C mismatches during sequencing, enabling the simultaneous profiling of total and labeled RNA expression from a single library.
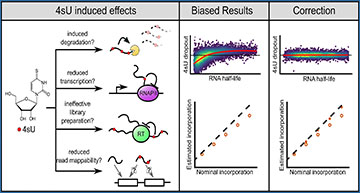
The researchers conducted experiments with escalating doses of 4sU and varying labeling durations in different cell lines. They observed that while short-term labeling (1h) had minimal impact, extended exposure to high concentrations of 4sU or longer labeling durations (up to 2h) led to biased expression estimates, particularly influenced by RNA half-life.
Further investigation revealed that this bias stemmed from multiple factors, including reduced mappability of reads carrying multiple nucleotide conversions and a global reduction in the representation of labeled RNA during library preparation. To address these challenges, the researchers developed innovative computational tools and statistical methods as part of the GRAND-SLAM pipeline and grandR package.
These tools not only rescued unmappable reads but also effectively mitigated remaining biases, offering a comprehensive solution for accurate RNA expression analysis in the context of nucleotide conversion RNA-seq.
Overall, this study sheds light on the dynamic nature of RNA synthesis and turnover, highlighting the importance of careful consideration when employing nucleoside analogues for labeling RNA. By unraveling the complexities of RNA dynamics, researchers can gain deeper insights into cellular processes and pave the way for future advancements in molecular biology research.
Berg K, Lodha M, Delazer I, Bartosik K, Garcia YC, Hennig T, Wolf E, Dölken L, Lusser A, Prusty BK, Erhard F. (2024) Correcting 4sU induced quantification bias in nucleotide conversion RNA-seq data. Nucleic Acids Res [Epub ahead of print]. [article]