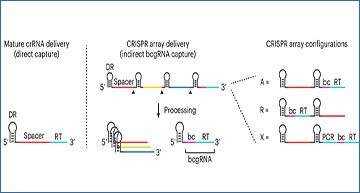
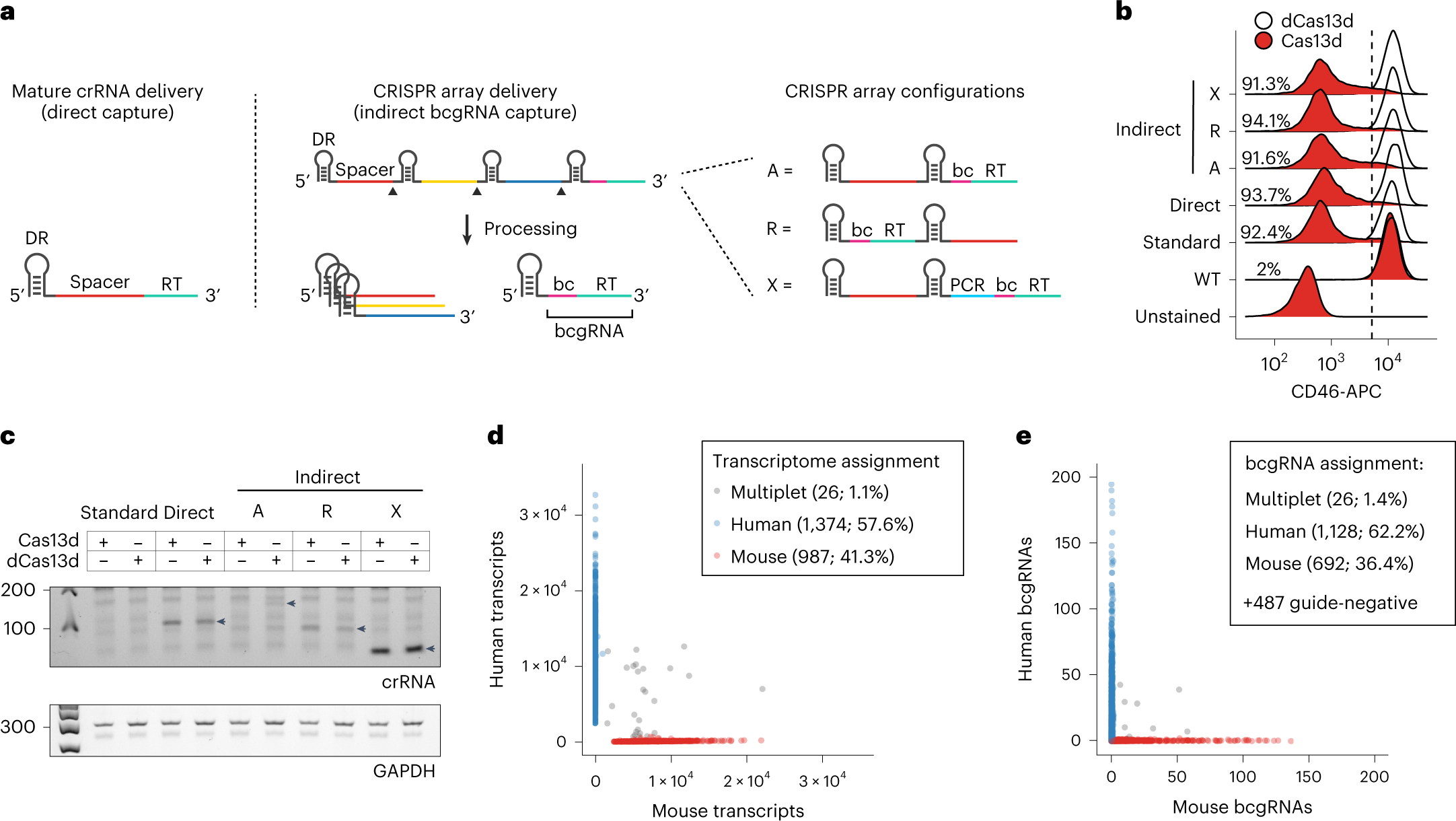
Pooled CRISPR screens coupled with single-cell RNA-sequencing have enabled systematic interrogation of gene function and regulatory networks. Researchers from the New York Genome Center have developed Cas13 RNA Perturb-seq (CaRPool-seq), which leverages the RNA-targeting CRISPR–Cas13d system and enables efficient combinatorial perturbations alongside multimodal single-cell profiling. CaRPool-seq encodes multiple perturbations on a cleavable CRISPR array that is associated with a detectable barcode sequence, allowing for the simultaneous targeting of multiple genes. The researchers compared CaRPool-seq to existing Cas9-based methods, highlighting its unique strength to efficiently profile combinatorially perturbed cells. Finally, they applied CaRPool-seq to perform multiplexed combinatorial perturbations of myeloid differentiation regulators in an acute myeloid leukemia (AML) model system and identify extensive interactions between different chromatin regulators that can enhance or suppress AML differentiation phenotypes./
Wessels HH, Méndez-Mancilla A, Hao Y, Papalexi E, Mauck WM 3rd, Lu L, Morris JA, Mimitou EP, Smibert P, Sanjana NE, Satija R. (2022) Efficient combinatorial targeting of RNA transcripts in single cells with Cas13 RNA Perturb-seq. Nat Methods [Epub ahead of print]. [abstract]