The success of mRNA vaccines has been realised, in part, by advances in manufacturing that enabled billions of doses to be produced at sufficient quality and safety. However, mRNA vaccines must be rigorously analysed to measure their integrity and detect contaminants that reduce their effectiveness and induce side-effects. Currently, mRNA vaccines and therapies are analysed using a range of time-consuming and costly methods. University of Queensland researchers have developed a streamlined method to analyse mRNA vaccines and therapies using long-read nanopore sequencing. Compared to other industry-standard techniques, VAX-seq can comprehensively measure key mRNA vaccine quality attributes, including sequence, length, integrity, and purity. The researchers also show how direct RNA sequencing can analyse mRNA chemistry, including the detection of nucleoside modifications. To support this approach, they provide supporting software to automatically report on mRNA and plasmid template quality and integrity. Given these advantages, these researchers anticipate that RNA sequencing methods, such as VAX-seq, will become central to the development and manufacture of mRNA drugs.
mRNA vaccine production and VAX-seq workflow
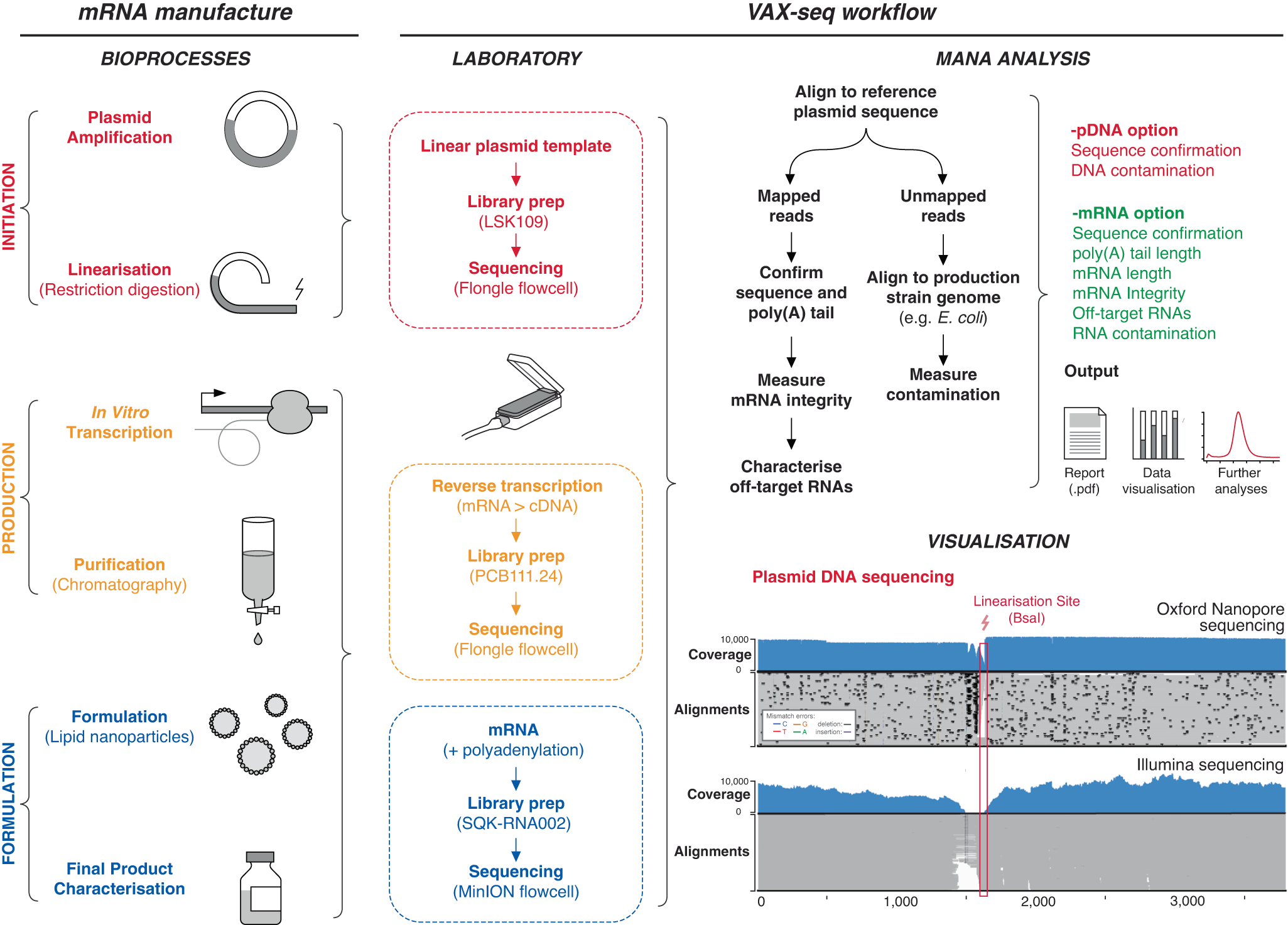
Schematic diagram illustrates the steps during mRNA manufacture (left panel), and the steps during VAX-seq analysis (right panel). This includes laboratory steps of long-read nanopore sequencing, followed by bioinformatic steps to analyse output data, including the supporting Mana software toolkit. mRNA vaccine quality features that can be analysed by VAX-seq are indicated (listed in red and green). In the bottom left corner is an IGV plot comparing Oxford Nanopore and Illumina sequencing of a plasmid DNA template. Coverage indicates the number of reads at each nucleotide position while the lower alignments grey bars indicate unique, individual alignments, with colouring indicating their similarity to the reference genome. Source data are provided as a Source data file.
Availability – Mana software is available at https://github.com/scchess/Mana
Gunter HM, Idrisoglu S, Singh S et al. (2023) mRNA vaccine quality analysis using RNA sequencing. Nat Commun 14, 5663 . [article]