RNA sequencing (RNA-seq) has revolutionized genomics by providing insights into gene expression patterns and regulatory mechanisms. However, despite its widespread use, RNA-seq analysis can be affected by reference bias, particularly in complex polyploid genomes. In this blog post, we delve into a recent study that sheds light on reference bias in RNA-seq analysis using hexaploid wheat as a model system.
Understanding Reference Bias
Reference bias occurs when RNA-seq quantification inaccurately measures transcripts derived from non-reference alleles, leading to erroneous downstream conclusions. This bias can be particularly pronounced in polyploid genomes, which contain multiple copies of each gene and may harbor introgressions from wild relatives.
The Study
Researchers at the Earlham Institute investigated reference bias in hexaploid wheat, a complex polyploid species known for its mosaic of wild relative introgressions. They used both simulated and experimental data to demonstrate widespread reference bias in RNA-seq alignment, primarily driven by divergent introgressed genes. This bias resulted in underestimated gene expression levels and misinterpretation of homoeologue expression balance.
Assessing the extent of reference bias in wheat
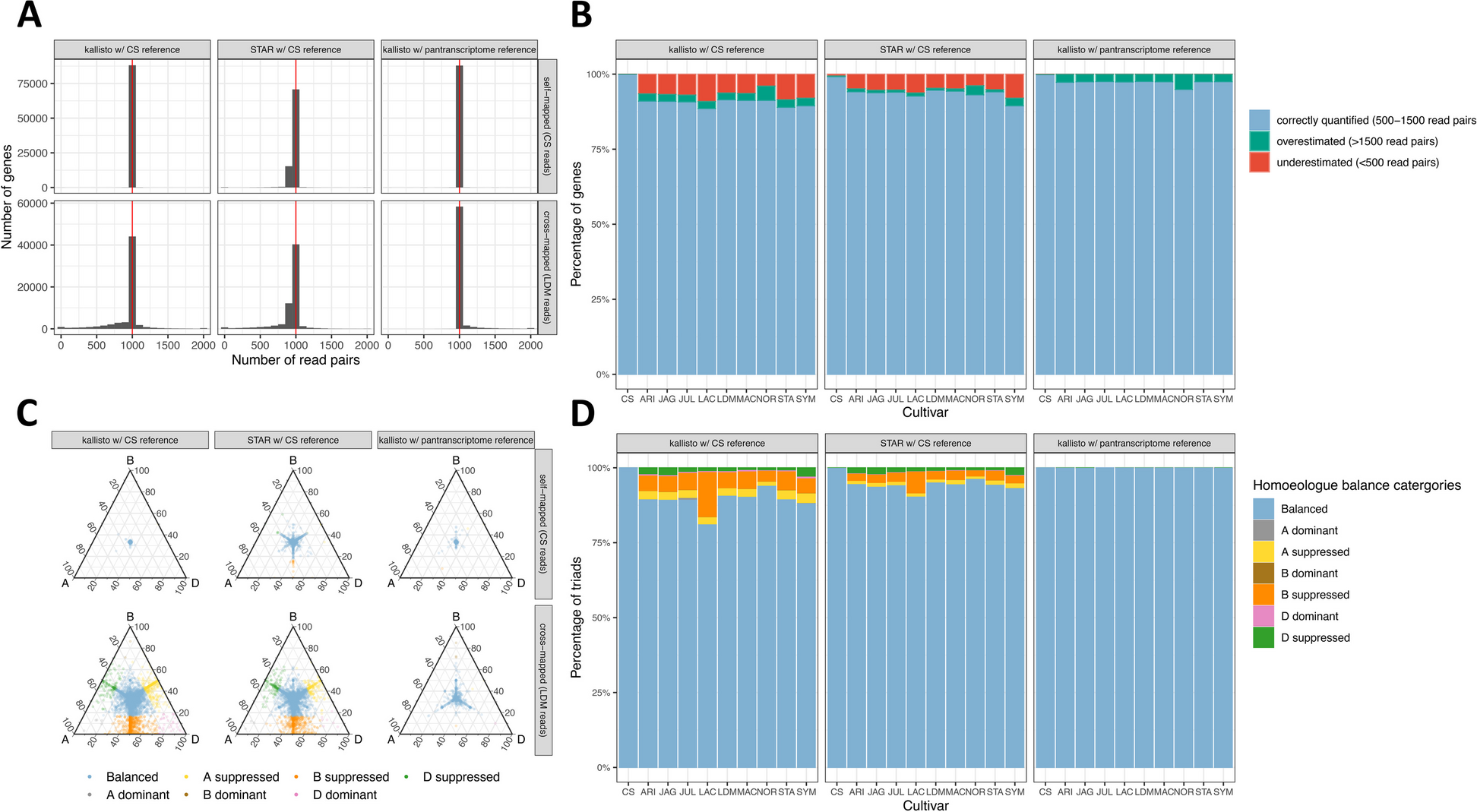
A Distribution of read counts when self-mapping Chinese Spring simulated reads or cross-mapping Landmark simulated reads. Comparing STAR and kallisto using the Chinese Spring RefSeq v1.0 reference and RefSeq v1.1 transcriptome and kallisto using the pantranscriptome reference. B Percentage of genes with expression estimated correctly, expression underestimated (< 500 read pairs) and expression overestimated (> 1500 read pairs) for simulated reads from 10 cultivars aligned to Chinese Spring with kallisto and STAR or to the pantranscriptome reference with kallisto. C Balance of homoeologue expression across triads when self-mapping Chinese Spring or cross-mapping Landmark simulated reads, comparing STAR and kallisto using the Chinese Spring RefSeq v1.0 reference and RefSeq v1.1 transcriptome and kallisto using the pantranscriptome reference. Each point on the ternary plot represents one triad. Points towards a corner indicate dominant expression of that homoeologue, and points opposite a corner indicate suppression of that homoeologue. D Percentage of triads in each expression category, using simulated reads from 10 cultivars aligned to Chinese Spring with kallisto and STAR or to the pantranscriptome reference with kallisto
Novel Method to Reduce Bias
To address this challenge, the researchers proposed a novel method to mitigate reference bias in wheat RNA-seq analysis. By incorporating gene models from multiple wheat genome assemblies into a pantranscriptome reference, they were able to reduce bias and capture a broader spectrum of genetic variation.
Implications and Future Directions
The study highlights the importance of considering reference bias in RNA-seq analysis, especially in complex polyploid genomes like wheat. Researchers using non-sample reference genomes for RNA-seq alignment should exercise caution and explore alternative methods to improve accuracy. The novel approach presented in this study offers a promising solution to mitigate reference bias and enhance the robustness of RNA-seq analysis in polyploid genomes.
Reference bias poses a significant challenge in RNA-seq analysis, particularly in complex polyploid genomes such as hexaploid wheat. By uncovering the extent of reference bias and proposing a novel method to address it, this study contributes valuable insights to the genomics community. Moving forward, researchers must remain vigilant about reference bias and continue to develop innovative strategies to ensure the accuracy and reliability of RNA-seq analysis in diverse biological contexts.
Coombes B, Lux T, Akhunov E, Hall A. (2024) Introgressions lead to reference bias in wheat RNA-seq analysis. BMC Biol 22(1):56. [article]