Nerve regeneration is a complex biological process that holds immense potential for clinical practice. While extensive research has been conducted on successful nerve regeneration, there remains a critical knowledge gap regarding failed regeneration, particularly in the formation of neuromas-in-continuity (NIC). To address this gap, a groundbreaking study has recently been conducted, employing a novel basic science model of rapid-stretch nerve injury combined with next-generation RNA sequencing. This study sheds light on the underlying cellular and molecular mechanisms that hinder successful nerve regeneration and provides valuable insights into the pathophysiology of NIC formation.
Unveiling the Transcriptional Landscape
University of Utah researchers applied next-generation RNA sequencing, a powerful technique that allows for the comprehensive analysis of the transcriptome, to investigate the temporal transcriptional landscape of pathophysiologic nerve regeneration. This cutting-edge approach provided a high-resolution snapshot of gene expression patterns throughout the regenerative process. By comparing the transcriptional profiles of NICs with those of successful nerve regeneration and Wallerian degeneration, the researchers were able to identify significant differences in genetic signatures.
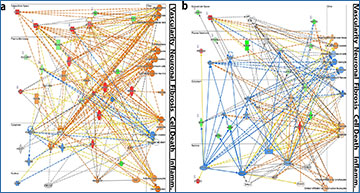
The results of this study revealed that pathophysiologic nerve regeneration leads to substantial alterations in genetic profiles, both temporally and in the mature neuroma microenvironment. These genetic signatures diverged from the coordinated patterns observed in traditional models of successful nerve regeneration. The analysis highlighted several prominent biological pathways that contribute to the development of NICs, including cellular death, fibrosis, neurodegeneration, metabolism, and unresolved inflammation.
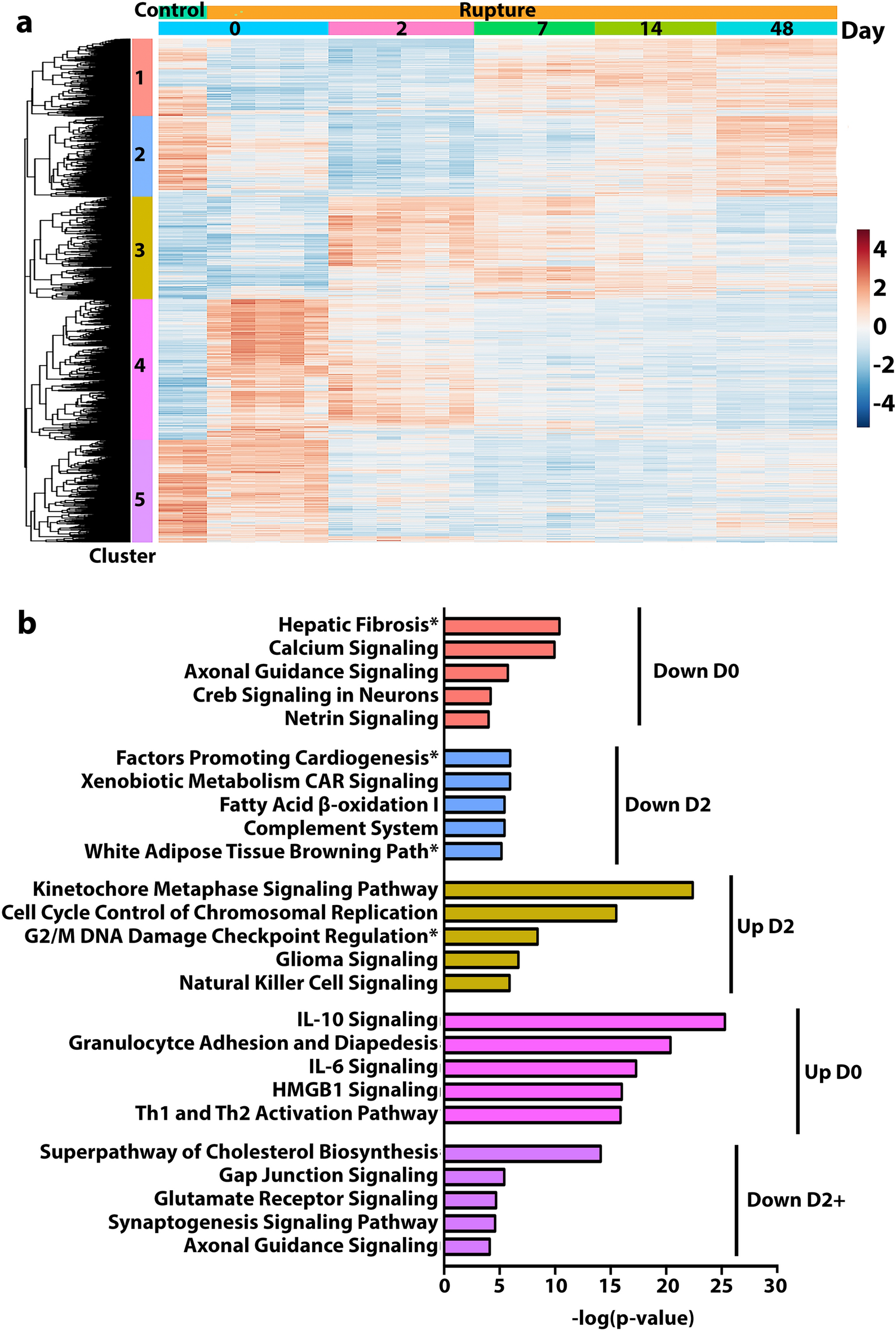
Cluster analysis to visualize the broad temporal patterns of genetic expression profiles with similar activity and application to IPA canonical pathways
(a) Cluster analysis and heatmap representation identified 5 clusters of genes demonstrating similar expression patterns across experimental timepoints. Clusters 1 and 4 reflect immediate changes in activity patterns, with Cluster 1 switching from up- to downregulation by 6 h after injury and Cluster 4 demonstrating the inverse pattern of down- to upregulation. Clusters 2, 3, and 5 depict delayed changes in activity; Clusters 2 and 5 express upregulated activity for control and then transition to downregulation by D2, while the inverse is observed for Cluster 3. Uniquely, Cluster 5 remains the most distinct from control expression levels at the terminal timepoint, while the other clusters return to similar expression profiles as their control counterparts. (b) Genes derived from the clustering were then subjected to IPA canonical pathways analysis to elucidate representative pathways, with the top 5 significant pathways presented. Down D0 (Cluster 1) genes mostly associate with fibrosis pathways and those implicated in neuronal specific activity. Metabolic regulation paths align with Down D2 (Cluster 2), whereas cell cycle regulation pathways are implicated with Up D2 (Cluster 3). Immune-related signatures dominate Up D0 (Cluster 4) whereas Down D2 + (Cluster 5) implicates pathways with both metabolic and neuronal underpinnings.
The application of next-generation RNA sequencing to investigate the transcriptional landscape of pathophysiologic nerve regeneration represents a groundbreaking milestone in our understanding of failed nerve regeneration and the formation of NICs. This study provides critical insights into the cellular and molecular underpinnings of NIC pathophysiology and highlights the divergent genetic signatures that distinguish unsuccessful nerve regeneration from successful regeneration. The identified biological pathways offer promising targets for future therapeutic interventions, bringing us closer to overcoming the challenges of failed nerve regeneration and improving patient outcomes. With further research and development, the knowledge gained from this study has the potential to revolutionize the field of nerve regeneration and transform clinical practice.
Warner WS, Stubben C, Yeoh S, Light AR, Mahan MA. (2023) Next-generation RNA sequencing elucidates transcriptomic signatures of pathophysiologic nerve regeneration. Sci Rep 13(1):8856. [article]