As an emerging sequencing technology, single-cell RNA sequencing (scRNA-Seq) has become a powerful tool for describing cell subpopulation classification and cell heterogeneity by achieving high-throughput and multidimensional analysis of individual cells and circumventing the shortcomings of traditional sequencing for detecting the average transcript level of cell populations. It has been applied to life science and medicine research fields such as tracking dynamic cell differentiation, revealing sensitive effector cells, and key molecular events of diseases. Researchers at Tianjin University discuss the recent technological innovations in scRNA-Seq, highlighting the latest research results with scRNA-Seq as the core technology in frontier research areas such as embryology, histology, oncology, and immunology. In addition, the researcher outline the prospects for its innovative application in traditional Chinese medicine (TCM) research and discusses the key issues currently being addressed by scRNA-Seq and its great potential for exploring disease diagnostic targets and uncovering drug therapeutic targets in combination with multiomics technologies.
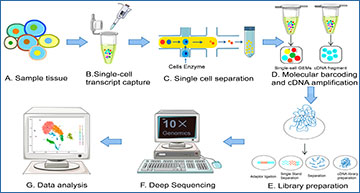
An overview of the single-cell RNA-sequencing procedures
(A) collect cells from tissue samples. (B) Single-cell capture process. (C) Cell isolation process. (D) Reverse transcription of mRNA and amplification of cDNA. (E) Construction of the scRNA-Seq library. (F) Complete deep sequencing of scRNA-Seq. (G) Analysis of scRNA-Seq data.
Wang S, Sun ST, Zhang XY, Ding HR, Yuan Y, He JJ, Wang MS, Yang B, Li YB. (2023) The Evolution of Single-Cell RNA Sequencing Technology and Application: Progress and Perspectives. Int J Mol Sci 24(3):2943. [article]