BACKGROUND
Just as single-cell sequencing has revolutionized many fields of biology, spatial “omics,” in which molecular parameters are measured in situ on intact tissue samples, is set to empower a new generation of scientific discoveries. A plethora of new technologies now enable spatial profiling of gene and protein expression, genetic mutations, epigenetic marks, chromatin structure, and genome organization. Most of these methods trace back to traditional techniques such as immunohistochemistry and in situ hybridization, or leverage the throughput of next-generation sequencing by converting spatial coordinates to sequence barcodes. Spatial omics technologies operate at vastly different levels of resolution, sensitivity, and throughput. There are also considerable differences in ease of adoption, compatibility with different sample types, commercial availability, upfront investment, and cost per sample. In such a rich and constantly evolving field, choosing the best technology to address a specific biological challenge—carefully weighting the strengths and weaknesses of each—is critical.
ADVANCES
The field of spatial molecular profiling has come of age over the past few years. Current RNA profiling technologies allow the spatial measurement of gene expression for centimeter-scale samples—in many cases, at single-cell resolution. Targeted methods can quantify thousands of transcripts, including those of very low abundance, whereas untargeted protocols offer transcriptome-wide profiling and can even detect gene isoforms and mutations, with coverage approaching that of disaggregated single-cell methods. Multiplexed immunohistochemistry protocols, either based on mass spectrometry or fluorescence imaging, can similarly be used to profile tens to hundreds of protein markers on a wide range of samples. Recent studies have reported additional approaches that recover other types of molecular information, including the spatial profiling of genomic organization for thousands of loci, open chromatin and epigenetic marks, and untargeted in situ DNA sequencing. Multi-omic technologies capturing multiple sources of information at once are also becoming available. As these methods become more reliable and widely adopted, it is becoming increasingly clear that spatial omics will contribute substantially to the elucidation of many biological questions, with prominent examples in fields ranging from oncology (such as the study of tumor heterogeneity and microenvironment) to neuroscience and organismal development.
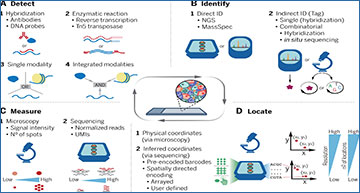
Spatial omics methods profile the molecular make-up of tissues, preserving their
spatial organization through four main steps
(A) Detection. Oligonucleotide probes or modified antibodies bind to specific nucleic acids, proteins, or small molecules. Alternatively, enzymatic processes can detect RNA (reverse transcription) or DNA (tagmentation) in an untargeted fashion. Single or multiple types of biomolecules can be profiled simultaneously. (B) Identification. Biomolecules can be identified directly with next-generation sequencing or mass spectrometry, or indirectly by means of an individual tag on the detection probe that is either read at once or sequentially (combinatorial barcode). (C) Measurement. Imaging-based methods measure the signal intensity of fluorescent probes or count the number of spots per area (single-molecule localization methods). Sequencing-based methods quantify the abundance of biomolecules through normalized read count or using unique molecular identifiers (UMIs). (D) Localization. Biomolecules are assigned to their spatial locations either directly in imaging-based methods, or by means of a DNA barcode decoded through sequencing. Technologies differ widely in terms of spatial resolution, sensitivity, and number and size of areas that can be profiled.
OUTLOOK
Future development trends in the spatial omic area are needed in three main areas: (i) multi-omics, which is defined as the simultaneous measurement of different parameters (for example, DNA, RNA, and protein); (ii) increased access and democratization, with technologies becoming more readily available, more reliable, and more robust; and (iii) improved analysis frameworks. As technologies continue to evolve, it will become necessary to dedicate more attention to data analysis and experimental design. Spatial omics technologies can already generate many terabytes of data in a single experiment, creating substantial challenges in data processing, analysis, and visualization. Statistical methods and guidelines for sample replication, study design, and batch correction are also lagging behind, although they’re evolving rapidly. Last, as the future keeps getting closer, we foresee that spatial omics will likely evolve toward three-dimensional spatial omics (operating on whole organs or even organisms) and spatial-temporal omics (performing measurements multiple times in vivo), adding further powerful and exciting tools to biology’s toolbox.
Bressan D, Battistoni G, Hannon GJ. (2023) The dawn of spatial omics. Science 381(6657):eabq4964. [abstract]