Understanding how genes are expressed within tissues and their spatial context is crucial for unraveling mysteries in health and disease. Imagine being able to pinpoint not only which genes are active but also where they are active within a tissue sample. This is where spatially resolved transcriptome (SRT) techniques come into play, offering a revolutionary approach that promises to transform various fields of biology.
Gene expression profiling, essentially studying which genes are turned on or off in a cell, has traditionally been done without considering the spatial arrangement of cells within tissues. However, with recent advancements in barcoding and sequencing chemistry, scientists can now explore the spatial dimension of gene expression. By correlating transcripts (molecules that carry genetic information) with their spatial locations within tissues, SRT techniques provide valuable insights into cell types, their locations, and even interactions between cells or molecules.
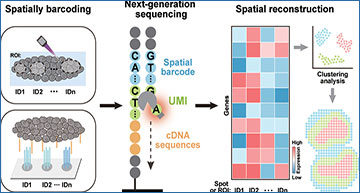
Schematic diagrams for workflows of next-generation sequencing (NGS)-based SRT methods, including spatial barcoding, library preparation, NGS, and spatial reconstruction
(A) Optical microdissection of ROIs followed by batch RNA-seq or batch scRNA-seq for SRT analysis. (B) Photochemical tagging of ROIs to spatially label cells or mRNAs followed by batch scRNA-seq/RNA-seq or bulk RNA-seq for SRT analysis. (C) DNA-barcoded array to spatially barcode cells or mRNAs at whole-tissue level followed by scRNA-seq or bulk RNA-seq for SRT analysis.
These techniques offer a dual advantage: not only do they provide information about the physical structure of tissues, but they also offer molecular insights, painting a comprehensive picture of the biological landscape. This has profound implications across several disciplines including developmental biology, neuroscience, oncology, and histopathology.
At the forefront of this exciting field are next-generation sequencing-based SRT methods, particularly those employing spatial barcoding chemistry. These methods utilize innovative approaches such as optically manipulated spatial indexing and DNA array-barcoded spatial indexing to precisely map gene expression within tissues.
Optically manipulated spatial indexing methods leverage sophisticated imaging techniques to tag specific locations within a tissue sample, allowing for precise mapping of gene expression. On the other hand, DNA array-barcoded spatial indexing methods utilize DNA barcodes attached to individual cells, enabling researchers to decode the spatial organization of gene expression.
However, like any emerging field, SRT techniques also face challenges. One major hurdle is the complexity of analyzing vast amounts of spatially resolved transcriptomic data. Developing robust computational tools capable of handling this data deluge is essential for realizing the full potential of SRT techniques.
Looking ahead, the future of spatially resolved transcriptomics holds immense promise. By overcoming current challenges and pushing the boundaries of technology, researchers aim to refine SRT techniques for even greater precision and scalability. Ultimately, these advancements will not only deepen our understanding of biological processes but also pave the way for novel diagnostics and therapeutics in the realms of health and disease.
Shi W, Zhang J, Huang S, Fan Q, Cao J, Zeng J, Wu L, Yang C. (2024) Next-Generation Sequencing-Based Spatial Transcriptomics: A Perspective from Barcoding Chemistry. JACS Au [Epub ahead of print]. [article]