Bladder cancer (BLCA) is one of the most common cancer types worldwide. The disease is responsible for about 200,000 deaths annually, thus improved diagnostics and therapy is needed. A large body of evidence reveal that small RNAs of less than 40 nucleotides may act as tumor suppressors, oncogenes, and disease biomarkers, with a major focus on microRNAs. However, the role of other families of small RNAs is not yet deciphered. Recent results suggest that small RNAs and their modification status, play a role in BLCA development and are promising biomarkers due to their high abundance in the exomes and body fluids (including urine). Moreover, free modified nucleosides have been detected at elevated levels from the urine of BLCA patients. A genome-wide view of small RNAs, and their modifications, will help pinpoint the molecules that could be used as biomarker or has important biology in BLCA development.
A team led by researchers at the University of Alabama at Birmingham obtained BLCA tumor tissue specimens from 12 patients undergoing transurethral resection of non-muscle invasive papillary urothelial carcinomas. They performed genome-wide profiling of small RNAs less than 40 bases long by a modified protocol with TGIRT (thermostable group II reverse transcriptase) to identify novel small RNAs and their modification status.
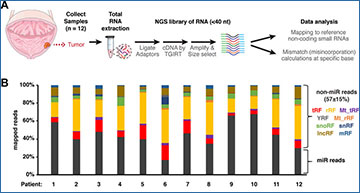
Comprehensive analysis identified not only microRNAs. Intriguingly, 57 ± 15% (mean ± S.D.) of sequencing reads mapped to non-microRNA-small RNAs including tRNA-derived fragments (tRFs), ribosomal RNA-derived fragments (rRFs) and YRNA-derived fragments (YRFs). Misincorporation (mismatch) sites identified potential base modification positions on the small RNAs, especially on tRFs, corresponding to m1A (N1-methyladenosine), m1G (N1-methylguanosine) and m2 2G (N2, N2-dimethylguanosine). The researchers also detected mismatch sites on rRFs corresponding to known modifications on 28 and 18S rRNA.
An updated small RNA-seq workflow for modification-friendly global analysis
(A) Scheme of collecting tumor samples from 12 NMIBC patients. Small RNA-sequencing libraries by TGIRT were prepared from total RNAs to profile relative abundance and potential RNA modifications (based on mismatch/misincorporation) for small RNAs less than 40 bases long. (B) Overall distribution of total mapped reads between microRNAs (dark grey) and non-microRNA small RNAs (nmsRNAs), including rRFs (yellow), tRFs (red) and more. (C) Distribution of mapped percentage for each sub-group of small RNAs shown as box-whisker plot. Box plot center represents median value, bounds represent upper and lower quartile, whiskers represent 1.5* interquartile range from the bounds.
These researchers found abundant non-microRNA-small RNAs in BLCA tumor samples. Small RNAs, especially tRFs and rRFs, contain modifications that can be captured as mismatch by TGIRT sequencing. Both the modifications and the non-microRNA-small RNAs should be explored as a biomarker for BLCA detection or follow-up.
Su Z, Monshaugen I, Klungland A, Ougland R, Dutta A. (2022) Characterization of novel small non-coding RNAs and their modifications in bladder cancer using an updated small RNA-seq workflow. Front Mol Biosci 9:887686. [article]