Long-read RNA sequencing (RNA-seq) is a powerful technology for transcriptome analysis, but the relatively low throughput of current long-read sequencing platforms limits transcript coverage. One strategy for overcoming this bottleneck is targeted long-read RNA-seq for preselected gene panels. Researchers at the Children’s Hospital of Philadelphia have developed TEQUILA-seq, a versatile, easy-to-implement, and low-cost method for targeted long-read RNA-seq utilizing isothermally linear-amplified capture probes. When performed on the Oxford nanopore platform with multiple gene panels of varying sizes, TEQUILA-seq consistently and substantially enriches transcript coverage while preserving transcript quantification. The researchers profile full-length transcript isoforms of 468 actionable cancer genes across 40 representative breast cancer cell lines. They identify transcript isoforms enriched in specific subtypes and discover novel transcript isoforms in extensively studied cancer genes such as TP53. Among cancer genes, tumor suppressor genes (TSGs) are significantly enriched for aberrant transcript isoforms targeted for degradation via mRNA nonsense-mediated decay, revealing a common RNA-associated mechanism for TSG inactivation. TEQUILA-seq reduces the per-reaction cost of targeted capture by 2-3 orders of magnitude, as compared to a standard commercial solution. TEQUILA-seq can be broadly used for targeted sequencing of full-length transcripts in diverse biomedical research settings.
Overview of TEQUILA-seq
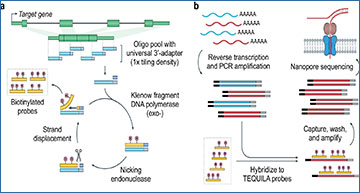
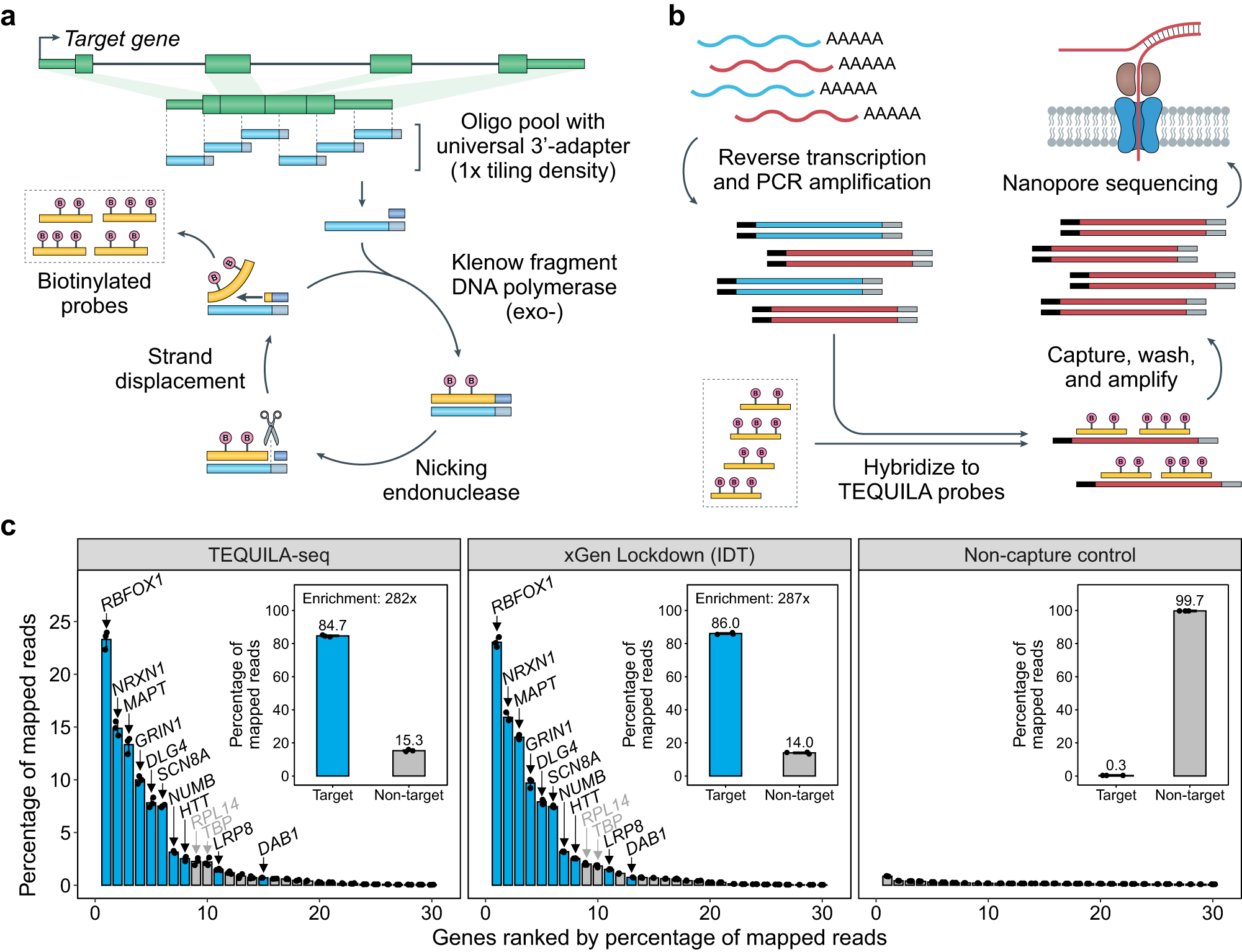
a, b Schematic of TEQUILA-seq. a Single-stranded DNA (ssDNA) oligonucleotides are designed to tile across all annotated exons of target genes and synthesized using an array-based DNA synthesis technology. TEQUILA probes are amplified from ssDNA oligo templates in a single pool using nickase-triggered strand displacement amplification with universal primers and biotin-dUTPs. b Full-length cDNAs are synthesized from poly(A)+ RNAs by reverse transcription and PCR amplification. TEQUILA probes are then hybridized to cDNAs. Upon capture and washing, cDNA-to-probe hybrids are immobilized to streptavidin magnetic beads, whereas unbound cDNAs are washed away. Captured cDNAs are further amplified by PCR and subjected to nanopore 1D library preparation and sequencing. c Comparison of TEQUILA-seq vs xGen Lockdown (IDT) probe-based target enrichment and sequencing. Main graphs: percentage of reads mapped to a given gene (mean ± s.d. of n = 3 replicates), for the top 30 genes with the highest number of mapped reads. Insets: percentage of reads mapped to target genes and non-target genes (mean ± s.d. of n = 3 replicates). Blue: target genes. Gray: non-target genes. Target gene panel: ten human genes with long transcripts in the brain. All sequencing methods were applied to the same human brain RNA mix from multiple donors (see Methods). Source data are provided as a Source Data file.
Availability – The scripts used for processing, analyzing, and visualizing TEQUILA-seq data are publicly available on GitHub (https://github.com/Xinglab/TEQUILA-seq) and deposited in Zenodo (https://doi.org/10.5281/zenodo.8018742).
Wang F, Xu Y, Wang R, Zhang B, Smith N, Notaro A, Gaerlan S, Kutschera E, Kadash-Edmondson KE, Xing Y, Lin L. (2023) TEQUILA-seq: a versatile and low-cost method for targeted long-read RNA sequencing. Nat Commun 14(1):4760. [article]