The accurate estimation of cell surface receptor abundance for single cell transcriptomics data is important for the tasks of cell type and phenotype categorization and cell-cell interaction quantification. Dartmouth College researchers previously developed an unsupervised receptor abundance estimation technique named SPECK (Surface Protein abundance Estimation using CKmeans-based clustered thresholding) to address the challenges associated with accurate abundance estimation. In that paper, the researchers concluded that SPECK results in improved concordance with Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) data relative to comparative unsupervised abundance estimation techniques using only single-cell RNA-sequencing (scRNA-seq) data.
In this paper, the researchers outline a new supervised receptor abundance estimation method called STREAK (gene Set Testing-based Receptor abundance Estimation using Adjusted distances and cKmeans thresholding) that leverages associations learned from joint scRNA-seq/CITE-seq training data and a thresholded gene set scoring mechanism to estimate receptor abundance for scRNA-seq target data. They evaluate STREAK relative to both unsupervised and supervised receptor abundance estimation techniques using two evaluation approaches on six joint scRNA-seq/CITE-seq datasets that represent four human and mouse tissue types. They conclude that STREAK outperforms other abundance estimation strategies and provides a more biologically interpretable and transparent statistical model.
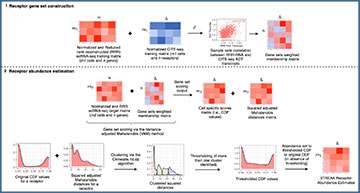
STREAK schematic with step (1) corresponding to the training co-expression analysis and step (2) corresponding to gene set scoring and subsequent clustering and thresholding to achieve cell-specific estimated receptor abundance profiles.
Javaid A, Frost HR (2023) STREAK: A supervised cell surface receptor abundance estimation strategy for single cell RNA-sequencing data using feature selection and thresholded gene set scoring. PLoS Comput Biol 19(8): e1011413. [article]