Alternative splicing (AS) is a crucial mechanism for regulating gene expression and isoform diversity in eukaryotes. However, the analysis and visualization of AS events from RNA sequencing data remains challenging. Most tools require a certain level of computer literacy and the available means of visualizing AS events, such as coverage and sashimi plots, have limitations and can be misleading. To address these issues, researchers at the University of Sydney have developed SpliceWiz, an R package with an interactive Shiny interface that allows easy and efficient AS analysis and visualization at scale. A novel normalization algorithm is implemented to aggregate splicing levels within sample groups, thereby allowing group differences in splicing levels to be accurately visualized. The tool also offers downstream gene ontology enrichment analysis, highlighting ASEs belonging to functional pathways of interest. SpliceWiz is optimized for speed and efficiency and introduces a new file format for coverage data storage that is more efficient than BigWig. Alignment files are processed orders of magnitude faster than other R-based AS analysis tools and on par with command-line tools. Overall, SpliceWiz streamlines AS analysis, enabling reliable identification of functionally relevant AS events for further characterization.
The SpliceWiz Pipeline
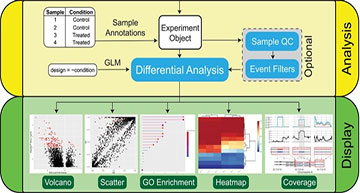
The SpliceWiz pipeline is organized into four modules. ‘Reference’ creates the SpliceWiz alternative splicing annotations required for processing alignments. ‘Experiment’ contains functions to process alignment files and collate their outputs into a dataset. ‘Analyse’ imports this dataset, allowing users to review quality control metrics and perform pre-test filtering of lowly-represented ASEs, and performs differential analysis to generate a table of results. ‘Display’ provides visualization tools, including volcano and scatter plots, gene ontology enrichment analysis, heatmaps and coverage plots.
Availability – SpliceWiz is a Bioconductor package available on GitHub (https://github.com/alexchwong/SpliceWiz)
Wong ACH, Wong JJ, Rasko JEJ, Schmitz U. (2023) SpliceWiz: interactive analysis and visualization of alternative splicing in R. Brief Bioinform 25(1):bbad468. [article]