Gene expression data is complex and may hold information regarding multiple biological processes at once. Researchers at Technion – Israel Institute of Technology have developed SPIRAL, an algorithm that uses a Gaussian statistical model to produce a comprehensive overview of a plurality of significant processes detected in single cell RNA-seq or spatial transcriptomics data. SPIRAL identifies biological processes by finding sub-matrices that consist of the subset of genes involved and the subset of cells or spots. We describe the algorithmic method, the analysis pipeline and several example results.
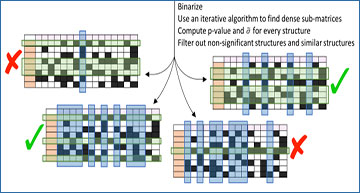
The SPIRAL algorithm pipeline
Structures marked with an X are filtered out; structures marked with a V are included in the reported structures. See text and Methods for details on the algorithm steps and the filtering criteria.
Availability – SPIRAL is available at: https://spiral.technion.ac.il/
Biran H, Hashimshony T, Mandel-Gutfreund Y, Yakhini Z. (2022) SPIRAL: Significant Process InfeRence ALgorithm for single cell RNA-sequencing and spatial transcriptomics. bioRXiv [online preprint]. [abstract]