With the emergence of single-cell RNA sequencing (scRNA-seq) technology, scientists are able to examine gene expression at single-cell resolution. Analysis of scRNA-seq data has its own challenges, which stem from its high dimensionality. The method of machine learning comes with the potential of gene (feature) selection from the high-dimensional scRNA-seq data. Even though there exist multiple machine learning methods that appear to be suitable for feature selection, such as penalized regression, there is no rigorous comparison of their performances across data sets, where each poses its own challenges.
Researchers at Thompson Rivers University analyzed and compared multiple penalized regression methods for scRNA-seq data. Given the scRNA-seq data sets they analyzed, the results show that sparse group lasso (SGL) outperforms the other six methods (ridge, lasso, elastic net, drop lasso, group lasso, and big lasso) using the metrics area under the receiver operating curve (AUC) and computation time. Building on these findings, the researchers proposed a new algorithm for feature selection using penalized regression methods. The proposed algorithm works by selecting a small subset of genes and applying SGL to select the differentially expressed genes in scRNA-seq data. By using hierarchical clustering to group genes, the proposed method bypasses the need for domain-specific knowledge for gene grouping information. In addition, the proposed algorithm provided consistently better AUC for the data sets used.
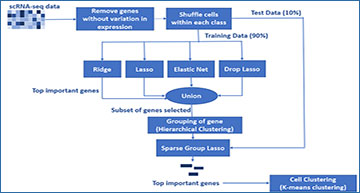
Schematic of the proposed algorithm for feature selection using penalized regression methods
There is a considerable reduction in the number of genes prior to the execution SGL. The top important genes selected by SGL are used to cluster cell groups using K-means clustering.
Puliparambil BS, Tomal JH, Yan Y. (2022) A Novel Algorithm for Feature Selection Using Penalized Regression with Applications to Single-Cell RNA Sequencing Data. Biology 11(10), 1495. [abstract]