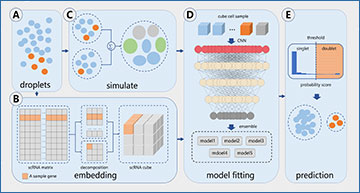
Doublets formed during single-cell RNA sequencing (scRNA-seq) severely affect downstream studies, such as differentially expressed gene analysis and cell trajectory inference, and limit the cellular throughput of scRNA-seq. Several doublet detection algorithms are currently available, but their generalization performance could be further improved due to the lack of effective feature-embedding strategies with suitable model architectures. Therefore, SoCube, a novel deep learning algorithm, was developed to precisely detect doublets in various types of scRNA-seq data. SoCube (i) proposed a novel 3D composite feature-embedding strategy that embedded latent gene information and (ii) constructed a multikernel, multichannel CNN-ensembled architecture in conjunction with the feature-embedding strategy. With its excellent performance on benchmark evaluation and several downstream tasks, it is expected to be a powerful algorithm to detect and remove doublets in scRNA-seq data.
SoCube workflow
Availability – SoCube is freely provided as an end-to-end tool on the Python official package site PyPi (https://pypi.org/project/socube/) and open-source on GitHub (https://github.com/idrblab/socube/).
Zhang H, Lu M, Lin G, Zheng L, Zhang W, Xu Z, Zhu F. (2023) SoCube: an innovative end-to-end doublet detection algorithm for analyzing scRNA-seq data. Brief Bioinform [Epub ahead of print]. [abstract]