Formalin-fixed paraffin-embedded (FFPE) tissues constitute a vast and valuable patient material bank for clinical history and follow-up data. It is still challenging to achieve single cell/nucleus RNA (sc/snRNA) profile in FFPE tissues. Researchers at Zhejiang University have developed a droplet-based snRNA sequencing technology (snRandom-seq) for FFPE tissues by capturing full-length total RNAs with random primers. snRandom-seq shows a minor doublet rate (0.3%), a much higher RNA coverage, and detects more non-coding RNAs and nascent RNAs, compared with state-of-art high-throughput scRNA-seq technologies. snRandom-seq detects a median of >3000 genes per nucleus and identifies 25 typical cell types. Moreover, the researchers applied snRandom-seq on a clinical FFPE human liver cancer specimen and revealed an interesting subpopulation of nuclei with high proliferative activity. This method provides a powerful snRNA-seq platform for clinical FFPE specimens and promises enormous applications in biomedical research.
snRandom-seq for FFPE tissues overview
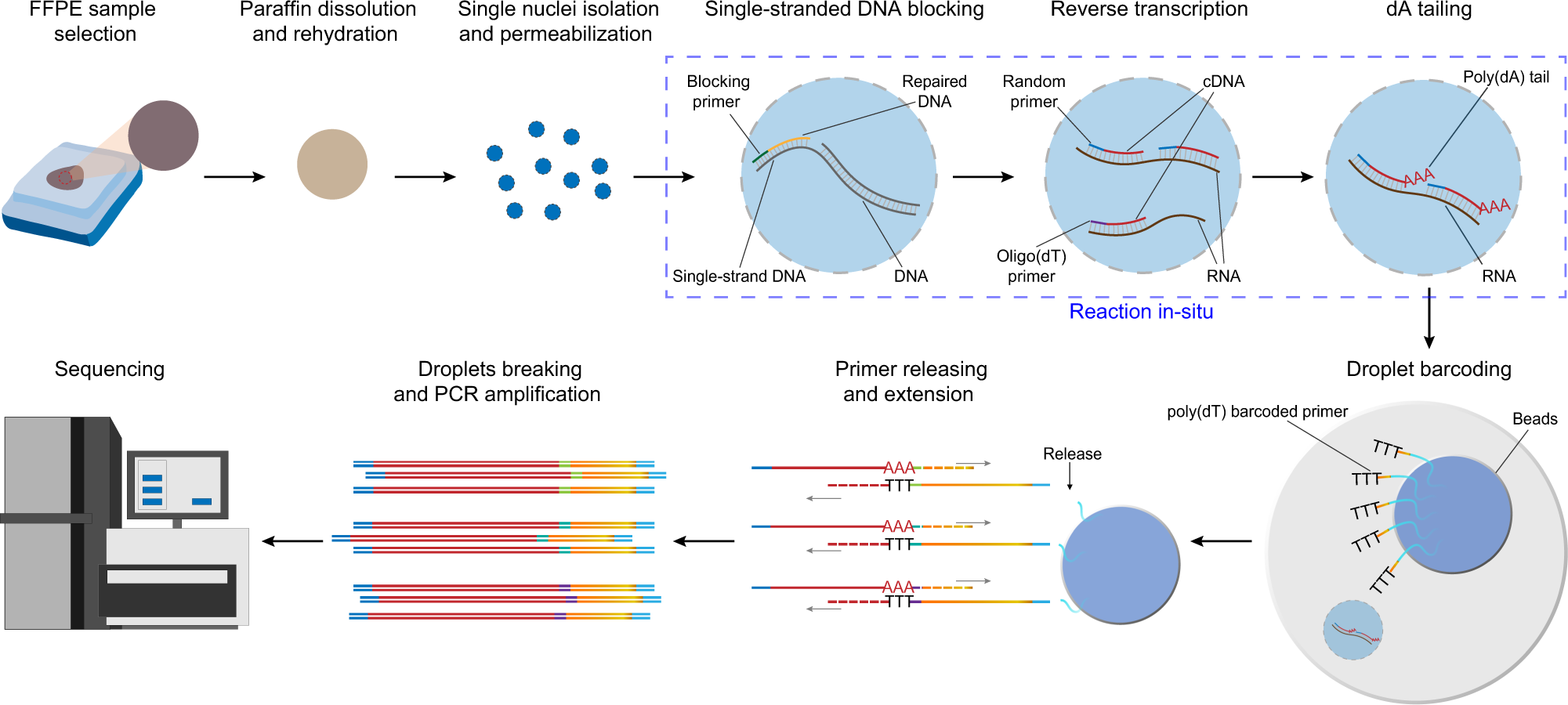
The workflow of snRandom-seq for FFPE tissues includes FFPE sample selection, paraffin dissolution, single nuclei isolation, and permeabilization, single-strand DNAs blocking, reverse transcription, dA tailing, droplet barcoding, primers releasing and extension, droplets breaking and PCR amplification, and sequencing. Red dashed circle in the FFPE tissue block: the areas of interest. Blue dashed box: the three in situ reactions, including single-strand DNAs blocking, reverse transcription, dA tailing. AAA: dA tail in the 3′ of cDNA. TTT: poly(dT) in the poly(dT) barcoded primers. Gray arrows: the direction of extension.
Xu Z, Zhang T, Chen H et al. (2023) High-throughput single nucleus total RNA sequencing of formalin-fixed paraffin-embedded tissues by snRandom-seq. Nat Commun 14, 2734. [article]