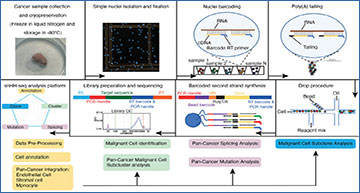
Tumor heterogeneity and its drivers impair tumor progression and cancer therapy. Single-cell RNA sequencing is used to investigate the heterogeneity of tumor ecosystems. However, most methods of scRNA-seq amplify the termini of polyadenylated transcripts, making it challenging to perform total RNA analysis and somatic mutation analysis.
Researchers at the Zhejiang University School of Medicine have developed a high-throughput and high-sensitivity method called snHH-seq, which combines random primers and a preindex strategy in the droplet microfluidic platform. This innovative method allows for the detection of total RNA in single nuclei from clinically frozen samples. A robust pipeline to facilitate the analysis of full-length RNA-seq data is also established. snHH-seq is applied to more than 730 000 single nuclei from 32 patients with various tumor types. The pan-cancer study enables it to comprehensively profile data on the tumor transcriptome, including expression levels, mutations, splicing patterns, clone dynamics, etc. New malignant cell subclusters and exploring their specific function across cancers are identified. Furthermore, the malignant status of epithelial cells is investigated among different cancer types with respect to mutation and splicing patterns. The ability to detect full-length RNA at the single-nucleus level provides a powerful tool for studying complex biological systems and has broad implications for understanding tumor pathology.
Workflow and evaluation of snHH-Seq
a) A schematic of the basic workflow for snHH-seq. b) Nuclei overloading boosts the percentage of droplets filled with nuclei. Nucleus concentration: ≈1000 000 nuclei mL−1, 4000 000 nuclei mL−1, 8000 000 nuclei mL−1. c) Expected collision rate as a function of the nuclei loading concentration for snHH-seq with different numbers of round1 barcodes. b,c) The line delineates the mean value, and the shading indicates the 95% confidence interval (CI). d) Scatter plot of human-mouse mix test using snHH-seq (48 preindex barcodes, load with ≈1000 000 nuclei mL−1, 50 µL droplet). Blue dots indicate human-specific cells; red dots indicate mouse-specific cells. Only 0.79% (purple dots) are human-mouse mixed cells. e) Saturation analysis of snHH-seq, 10X Chromium V3 and VASA-drop. The number of genes detected per nucleus when downsampling total read counts to the indicated depths. f) Read coverage along the gene body for snHH-seq, 10X Chromium V3, and VASA-drop.
Chen H, Fang X, Shao J, Zhang Q, Xu L, Chen J, Mei Y, Jiang M, Wang Y, Li Z, Chen Z, Chen Y, Yu C, Ma L, Zhang P, Zhang T, Liao Y, Lv Y, Wang X, Yang L, Fu Y, Chen D, Jiang L, Yan F, Lu W, Chen G, Shen H, Wang J, Wang C, Liang T, Han X, Wang Y, Guo G. (2023) Pan-Cancer Single-Nucleus Total RNA Sequencing Using snHH-Seq. Adv Sci (Weinh) [Epub ahead of print]. [article]