Single-cell RNA-sequencing is revolutionising the study of cellular and tissue-wide heterogeneity in a large number of biological scenarios, from highly tissue-specific studies of disease to human-wide cell atlases. A central task in single-cell RNA-sequencing analysis design is the calculation of cell type-specific genes in order to study the differential impact of different replicates (e.g. tumour vs. non-tumour environment) on the regulation of those genes and their associated networks. The crucial task is the efficient and reliable calculation of such cell type-specific ‘marker’ genes. These optimise the ability of the experiment to isolate highly-specific cell phenotypes of interest to the analyser. However, while methods exist that can calculate marker genes from single-cell RNA-sequencing, no such method places emphasise on specific cell phenotypes for downstream study in e.g. differential gene expression or other experimental protocols (spatial transcriptomics protocols for example). Researchers from the European Bioinformatics Institute have developed SMaSH, a general computational framework for extracting key marker genes from single-cell RNA-sequencing data which reliably characterise highly-specific and niche populations of cells in numerous different biological data-sets.
SMaSH extracts robust and biologically well-motivated marker genes, which characterise a given single-cell RNA-sequencing data-set better than existing computational approaches for general marker gene calculation. The researchers demonstrate the utility of SMaSH through its substantial performance improvement over several existing methods in the field. Furthermore, they evaluate the SMaSH markers on spatial transcriptomics data, demonstrating they identify highly localised compartments of the mouse cortex.
The SMaSH framework
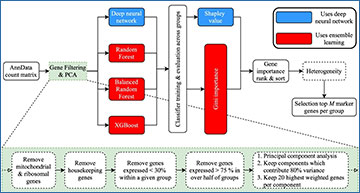
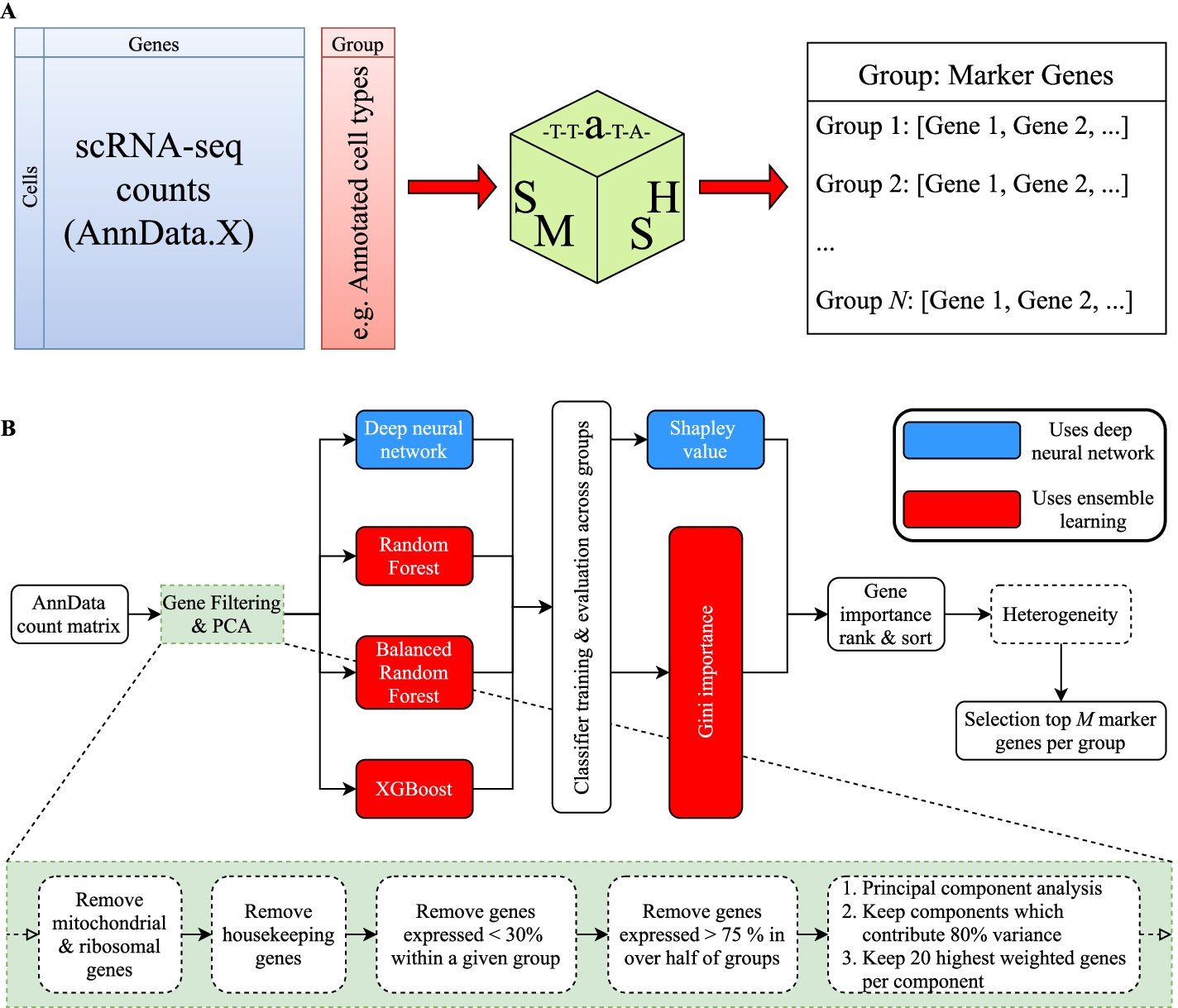
A SMaSH works directly from the counts matrix, producing a dictionary relating the user-defined classes of interest (e.g. cell type annotations) to top marker genes for each class (default top 5). B SMaSH filters and ranks genes according to an ensemble learning model or a deep neural network
SMaSH is a new methodology for calculating robust markers genes from large single-cell RNA-sequencing data-sets, and has implications for e.g. effective gene identification for probe design in downstream analyses spatial transcriptomics experiments. SMaSH has been fully-integrated with the ScanPy framework and provides a valuable bioinformatics tool for cell type characterisation and validation in every-growing data-sets spanning over 50 different cell types across hundreds of thousands of cells.
Availability – The complete SMaSH implementation, including several full examples of how to use SMaSH and reproduce the results in the paper are available under the Cvejic group GitLab: https://gitlab.com/cvejic-group/smash
Nelson ME, Riva SG, Cvejic A. (2022) SMaSH: a scalable, general marker gene identification framework for single-cell RNA-sequencing. BMC Bioinformatics 23(1):328. [article]